Visualisation and Plotting¶
Fleur specific Plotting¶
Plotting routine for fleur density of states and bandstructures
-
masci_tools.vis.fleur.plot_fleur_bands(bandsdata, bandsattributes, spinpol=True, bokeh_plot=False, weight=None, **kwargs)[source]¶ Plot the bandstructure previously extracted from a banddos.hdf via the
HDF5ReaderThis routine expects datasets and attributes read in with the FleurBands recipe from
recipesor something producing equivalent data- Parameters
dosdata – dataset dict produced by the FleurBands recipe
attributes – attributes dict produced by the FleurBands recipe
spinpol – bool, if True (default) use the plot for spin-polarized bands if the data is spin-polarized
bokeh_plot – bool (default False), if True use the bokeh routines for plotting
weight – str, name of the weight (without spin suffix _up or _dn) you want to emphasize
- All other Kwargs are passed on to the underlying plot routines
Matplotlib:
plot_bands(),plot_spinpol_bands()Bokeh:
bokeh_bands(),bokeh_spinpol_bands()
-
masci_tools.vis.fleur.plot_fleur_dos(dosdata, attributes, spinpol=True, bokeh_plot=False, multiply_by_equiv_atoms=False, plot_keys=None, show_total=True, show_interstitial=True, show_sym=False, show_atoms='all', show_lresolved=None, key_mask=None, **kwargs)[source]¶ Plot the density of states previously extracted from a banddos.hdf via the
HDF5ReaderThis routine expects datasets and attributes read in with the FleurDOS (Or related DOS modes) recipe from
recipesor something producing equivalent data- Parameters
dosdata – dataset dict produced by the FleurDOS recipe
attributes – attributes dict produced by the FleurDOS recipe
spinpol – bool, if True (default) use the plot for spin-polarized dos if the data is spin-polarized
bokeh_plot – bool (default False), if True use the bokeh routines for plotting
- Arguments for selecting the DOS components to plot:
- param plot_keys
optional str list of str, defines the labels you want to plot
- param show_total
bool, if True (default) the total DOS is shown
- param show_interstitial
bool, if True (default) the interstitial DOS is shown
- param show_atoms
either ‘all’, None, or int or list of ints, defines, which total atom projections to show
- param show_atoms
either ‘all’, None, or int or list of ints, defines, which total atom projections to show
- param key_mask
list of bools of the same length as the number of datasets, alternative way to specify, which entries to plot
- All other Kwargs are passed on to the underlying plot routines
Matplotlib:
plot_dos(),plot_spinpol_dos()Bokeh:
bokeh_dos(),bokeh_spinpol_dos()
KKR specific Plotting¶
-
masci_tools.vis.kkr_plot_FS_qdos.FSqdos2D(p0='./', totonly=True, s=20, ls_ef=':', lw_ef=1, color='', reload_data=False, clrbar=True, atoms=[], ax=None, nosave=False, noalat=False, cmap=<matplotlib.colors.LinearSegmentedColormap object>, noplot=False, return_data=False, pclrmesh=False, logscale=True, ef=None)[source]¶ plotting routine for dos files
dispersionplot function for plotting KKR bandstructures (i.e. qdos) files
-
masci_tools.vis.kkr_plot_bandstruc_qdos.dispersionplot(p0='./', totonly=True, s=20, ls_ef=':', lw_ef=1, units='eV_rel', noefline=False, color='', reload_data=False, clrbar=True, logscale=True, nosave=False, atoms=None, ratios=False, atoms2=None, noscale=False, newfig=False, cmap=None, alpha=1.0, qcomponent=- 2, clims=None, xscale=1.0, raster=True, atoms3=None, alpha_reverse=False, return_data=False, xshift=0.0, yshift=0.0, plotmode='pcolor', ptitle=None, ef=None, as_e_dimension=None, scale_alpha_data=False, shading='gouraud')[source]¶ plotting routine for qdos files - dispersion (E vs. q)
-
masci_tools.vis.kkr_plot_dos.dosplot(p0='./', totonly=True, color='', label='', marker='', lw=2, ms=5, ls='-', ls_ef=':', lw_ef=1, units='Ry', noefline=False, interpol=False, allatoms=False, onespin=False, atoms=[], lmdos=False, lm=[], nofig=False, scale=1.0, shift=0, normalized=False, xyswitch=False, efcolor='', return_data=False, xscale=1.0, xshift=0.0, yshift=0.0, filled=False, spins=2)[source]¶ plotting routine for dos files
-
masci_tools.vis.kkr_plot_shapefun.change_zoom(ax, zoom_range, center=[0, 0, 0])[source]¶ Change the zoom of a 3d plot
- Author
Philipp Ruessmann
- Parameters
ax – axis which is zoomed
zoom_range – range to which the image is zoomed, total range from center-zoom_range to center+zoom_range
center – center of the zoomed region (optional, defaults to origin)
-
masci_tools.vis.kkr_plot_shapefun.plot_shapefun(pos, out, mode)[source]¶ Creates a simple matplotlib image to show the shapefunctions given it’s positions in the unit cell, the atoms’s vertices in ut and the plotting mode
- Author
Philipp Ruessmann
- Parameters
pos – positions of the centers of the cells
verts – array of vertices of the shapefunction (outlines of shapes)
mode – ‘all’ or ‘single’ determines whether or not all shapes are combined in a single figure or plotted as individual figures
- Returns ax
return the axis in which the plot was done (useful to pass to ‘change_zoom’ and ‘zoom_in’ functions of this module
-
masci_tools.vis.kkr_plot_shapefun.zoom_in(ax, atm, pos, zoom_range=10)[source]¶ Zoom into shapefun of a single atom
- Author
Philipp Ruessmann
- Parameters
ax – axis in which shapefun plot is found
atm – atom index whose shapefunction is zoomed
pos – array of positions of centers of the shapes (needed to shift center of zommed region to correct atom
zoom_range – range of the zoomed region (optional, defaults to 10)
General Plotting¶
Here basic functionality is provided for setting default parameters for plotting and ensuring consitent values for these
-
masci_tools.vis.NestedPlotParameters(plotter_object)[source]¶ Contextmanager for nested plot function calls Will reset function defaults and parameters to previous values after exiting
- Parameters
plotter_object – Plotter instance
-
class
masci_tools.vis.Plotter(default_parameters, general_keys=None, key_descriptions=None, **kwargs)[source]¶ Base class for handling parameters for plotting methods. For different plotting backends a subclass can be created to represent the specific parameters of the backend.
- Args:
- param default_parameters
dict with hardcoded default parameters
- param general_keys
set of str optional, defines parameters which are not allowed to change for each entry in the plot data
Kwargs in the __init__ method are forwarded to
Plotter.set_defaults()to change the current defaults away from the hardcoded parameters.The Plotter class creates a hierachy of dictionaries for lookups on this object utilizing the ChainMap from the collections module.
- The hierachy is as follows (First entries take precedence over later entries):
parameters: set by
set_parameters()(usually arguments passed into function)user defaults: set by
set_defaults()function defaults: set by
set_defaults()with default_type=’function’global defaults: Hardcoded as fallback
Only the parameters can represent parameters for multiple sets of plot calls. All others are used as fallback for specifying non-specified values for single plots
The current parameters can be accessed by bracket indexing the class. A example of this is shown below.
parameter_dict = {'fontsize': 16, 'linestyle': '-'} params = Plotter(parameter_dict) #Accessing a parameter print(params['fontsize']) # 16 #Modifying a parameter params['fontsize'] = 20 print(params['fontsize']) # 20 #Creating a parameter set for multiple plots #1. Set the properties to the correct values params.single_plot = False params.num_plots = 3 #2. Now we can set a property either by providing a list or a integer indexed dict # Both of the following examples set the linestyle of the second and third plot to '--' params['linestyle'] = [None, '--', '--'] params['linestyle'] = {1: '--', 2: '--'} # Not specified values are replaced with the default value for a single plot print(params['linestyle']) # ['-', '--', '--'] #In lists properties can also be indexed via tuples print(params[('linestyle', 0)]) # '-' print(params[('linestyle', 1)]) # '--' #Changes to the parameters and properties are reset params.reset_parameters() print(params['linestyle']) # '-'
-
add_parameter(name, default_from=None, default_val=None)[source]¶ Add a new parameter to the parameters dictionary.
- Parameters
name – str name of the parameter
default_from – str (optional), if given a entry is created in the curent defaults with the name and the default value of the key default_from
-
static
convert_to_complete_list(given_value, single_plot, num_plots, default=None, key='')[source]¶ Converts given value to list with length num_plots with None for the non-specified values
- Parameters
given_value – value passed in, for multiple plots either list or dict with integer keys
single_plot – bool, if True only a single parameter is allowed
num_plots – int, if single_plot is False this defines the number of plots
default – default value for unspecified entries
key – str of the key to process
-
static
dict_of_lists_to_list_of_dicts(dict_of_lists, single_plot, num_plots)[source]¶ Converts dict of lists and single values to list of length num_plots or single dict for single_plot=True
- Parameters
dict_of_lists – dict to be converted
single_plot – boolean, if True only a single parameter set is allowed
num_plots – int of the number of allowed plots
- Returns
list of dicts
-
get_description(key)[source]¶ Get the description of the given key
- Parameters
key – str of the key, for which the description should be printed
-
get_multiple_kwargs(keys, ignore=None)[source]¶ Get multiple parameters and return them in a dictionary
- Parameters
keys – set of keys to process
ignore – str or list of str (optional), defines keys to ignore in the creation of the dict
-
property
num_plots¶ Integer property for number of plots produced
-
remove_added_parameters()[source]¶ Remove the parameters added via
Plotter.add_parameter()
-
reset_defaults()[source]¶ Resets the defaults to the hardcoded defaults in _PLOT_DEFAULTS. Will check beforehand if the parameters or properties differ from the defaults and will raise an error if this is the case
-
reset_parameters()[source]¶ Reset the parameters to the current defaults. The properties single_plot and num_plots are also set to default values
-
set_defaults(continue_on_error=False, default_type='global', **kwargs)[source]¶ Set the current defaults. This method will only work if the parameters are not changed from the defaults. Otherwise a error is raised. This is because after changing the defaults the changes will be propagated to the parameters to ensure consistency.
- Parameters
continue_on_error – bool, if True unknown key are simply skipped
- Default_type
either ‘global’ or ‘function’. Specifies, whether to set the global defaults (not reset after function) or the function defaults
Kwargs are used to set the defaults.
-
set_parameters(continue_on_error=False, **kwargs)[source]¶ Set the current parameters.
- Parameters
continue_on_error – bool, if True unknown key are simply skipped and returned
Kwargs are used to set the defaults.
-
set_single_default(key, value, default_type='global')[source]¶ Set default value for a single key/value pair
- Parameters
key – str of the key to set
value – value to set the key to
- Default_type
either ‘global’ or ‘function’. Specifies, whether to set the global defaults (not reset after function) or the function defaults
-
property
single_plot¶ Boolean property if True only a single Plot parameter set is allowed
-
masci_tools.vis.ensure_plotter_consistency(plotter_object)[source]¶ Decorator for plot functions to ensure that the Parameters are reset even if an error occurs in the function Additionally checks are performed that the parameters are reset after execution and the defaults are never changed in a plot function
- Parameters
plotter_object – Plotter instance to be checked for consistency
Matplotlib¶
This module contains a subclass of Plotter for the matplotlib library
-
class
masci_tools.vis.matplotlib_plotter.MatplotlibPlotter(**kwargs)[source]¶ Class for plotting parameters and standard code snippets for plotting with the matplotlib backend.
Kwargs in the __init__ method are forwarded to setting default values for the instance
For specific documentation about the parameter/defaults handling refer to
Plotter.Below the current defined default values are shown:
Plot Parameters¶ Name
Description
Default value
title_fontsizeFontsize for the title of the figure
16figure_kwargsArguments passed to plt.figure when creating the figure. Includes things like figsize, dpi, background color, …
{‘figsize’: (8, 6), ‘dpi’: 600, ‘facecolor’: ‘w’, ‘edgecolor’: ‘k’, ‘constrained_layout’: False}
alphaFloat specifying the transparency of the title
1axis_linewidthLinewidth of the lines for the axis
1.5use_axis_formatterIf True the labels will always not be formatted with an additive constant at the top
Falseset_powerlimitsIf True the threshold for switching to scientific notation is adjusted to 0,3
TruexticksPositions of the ticks on the x axis
No Default
xticklabelsLabels for the ticks on the x-axis
No Default
yticksPositions for the ticks on the y-axis
No Default
yticklabelsLabels for the ticks on the y-axis
No Default
invert_xaxisIf True the direction of the x-axis is inverted
Falseinvert_yaxisIf True the direction of the y-axis is inverted
Falsecolor_cycleIf set this will override the default color cycle of matplotlib. Can be given as name of a colormap cycle or list of colors
No Default
sub_colormapIf a colormap is used this can be used to cut out a part of the colormap. For example (0.5,1.0) will only use the upper half of the colormap
No Default
linewidthLinewidth for the plot(s)
2.0linestyleLinestyle for the plot(s)
-markerShape of the marker to use for the plot(s)
omarkersizeSize of the markers to use in the plot(s)
4.0colorColor to use in the plot(s)
No Default
zorderz-position to use for the plot(s) (Is used to define fore- and background)
No Default
repeat_colors_afterIf set the colors will be repeated after the given number of plots. Only implemented for multiple_scatterplots
No Default
edgecolorEdgecolor to use in the plot(s)
No Default
facecolorFacecolor to use in the plot(s)
No Default
plot_labelLabel to use in the plot(s) for the legend
No Default
area_plotIf True fill_between(x) will be used to produce the plot(s)
Falsearea_verticalDetermines, whether to use fill_between or fill_betweenx for area plots
Falsearea_enclosing_lineIf True a enclosing line will be drawn around the area
Truearea_alphaTransparency to use for the area in the area plot(s)
1.0area_linecolorColor for the enclosing line in the area plot(s)
No Default
plot_alphaTransparency to use for the plot(s)
1.0cmapColormap to use for scatter/pcolormesh or 3D plots
viridisnormIf set this norm will be used to normalize data for the colormapping
No Default
shadingShading to use for pcolormesh plots
gouraudrasterizedRasterize the pcolormesh when drawing vector graphics.
TruescaleDict specifying the scales of the axis, e.g {‘y’: ‘log’}will create a logarithmic scale on the y-axis
No Default
limitsDict specifying the limits of the axis, e.g {‘x’: (-5,5)}
No Default
labelfontsizeFontsize for the labels on the axis
15linesDict specifying straight help-lines to draw. For example {‘vertical’: 0, ‘horizontal’: [-1,1]} will draw a vertical line at 0 and two horizontal at -1 and 1
No Default
line_optionsColor, width, and more options for the help-lines
{‘linestyle’: ‘–’, ‘color’: ‘k’, ‘linewidth’: 1.0}
font_optionsDefault font options that can be used for text annotations
{‘family’: ‘serif’, ‘color’: ‘black’, ‘weight’: ‘normal’, ‘size’: 16}
tick_paramsxParameters for major ticks on the x-axis (Size, fontsize, …)
{‘size’: 4.0, ‘width’: 1.0, ‘labelsize’: 14, ‘length’: 5, ‘labelrotation’: 0}
tick_paramsyParameters for major ticks on the y-axis (Size, fontsize, …)
{‘size’: 4.0, ‘width’: 1.0, ‘labelsize’: 14, ‘length’: 5, ‘labelrotation’: 0}
tick_paramsx_minorParameters for minor ticks on the x-axis (Size, fontsize, …)
{‘size’: 2.0, ‘width’: 1.0, ‘labelsize’: 0, ‘length’: 2.5}
tick_paramsy_minorParameters for minor ticks on the y-axis (Size, fontsize, …)
{‘size’: 2.0, ‘width’: 1.0, ‘labelsize’: 0, ‘length’: 2.5}
colorbarIf True and the function implements color mapping, a colorbar is shown
Truecolorbar_paddingSpecifies the space between plot and colorbar
0.1legendIf True a legend for the plot is shown
Falselegend_optionsParameters for displaying the legend (Fontsize, location, …)
{‘fontsize’: 16, ‘linewidth’: 3.0, ‘loc’: ‘best’, ‘fancybox’: True}
save_plotsif True the plots will be saved to file
Falsesave_formatFormats to save the plots to, can be single or list of formats
pngsave_optionsAdditional options for saving the plots to file
{‘transparent’: True}
tightlayoutIf True the tight layout will be used (NOT IMPLEMENTED)
FalseshowIf True plt.show will be called at the end of the routine
Truesave_raw_plot_dataIf True the data for the plot is saved to file (NOT IMPLEMENTED)
Falseraw_plot_data_formatFormat in which to save the data for the plot (NOT IMPLEMENTED)
txt-
draw_lines(ax)[source]¶ Draw horizontal and vertical lines specified in the lines argument
- Parameters
ax – Axes object on which to perform the operation
-
plot_kwargs(ignore=None, extra_keys=None, plot_type='default', post_process=True, **kwargs)[source]¶ Creates a dict or list of dicts (for multiple plots) with the defined parameters for the plotting calls fo matplotlib
- Parameters
ignore – str or list of str (optional), defines keys to ignore in the creation of the dict
extra_keys – optional set for addtional keys to retrieve
post_process – bool, if True the parameters are cleaned up for inserting them directly into matplotlib plitting functions
Kwargs are used to replace values by custom parameters:
Example for using a custom markersize:
p = MatplotlibPlotter() p.add_parameter('marker_custom', default_from='marker') p.plot_kwargs(marker='marker_custom')
This code snippet will return the standard parameters for a plot, but the value for the marker will be taken from the key marker_custom
-
prepare_plot(title=None, xlabel=None, ylabel=None, zlabel=None, axis=None, minor=False, projection=None)[source]¶ Prepares the figure of a matplotlib plot, setting the labels/titles, ticks, …
- Parameters
title – str for the title of the figure
xlabel – str for the label on the x-axis
ylabel – str for the label on the y-axis
zlabel – str for the label on the z-axis
axis – matplotlib axes object, optional, if given the operations are performed on the object otherwise a new figure and subplot are created
minor – bool, if True minor tick parameters are set
projection – str, passed on to the add_subplot call
- Returns
the created or modified axis object
-
save_plot(saveas)[source]¶ Save the current figure or show the current figure
- Parameters
saveas – str, filename for the resulting file
-
set_limits(ax)[source]¶ Set limits of the axis
- Parameters
ax – Axes object on which to perform the operation
-
set_scale(ax)[source]¶ Set scale of the axis (for example ‘log’)
- Parameters
ax – Axes object on which to perform the operation
-
show_colorbar(ax)[source]¶ Print a colorbar for the plot
- Parameters
ax – Axes object on which to perform the operation
-
show_legend(ax, leg_elems=None)[source]¶ Print a legend for the plot
- Parameters
ax – Axes object on which to perform the operation
-
static
truncate_colormap(cmap, minval=0.0, maxval=1.0, n=256)[source]¶ Cut off parts of colormap
- Parameters
cmap – cmap to truncate
minval – minimum value of new colormap
maxval – maximum value of new colormap
n – number of colors in new colormap
- Returns
colormap truncated to only hold colors between minval and maxval from old colormap
-
In this module are plot routines collected to create default plots out of certain ouput nodes from certain workflows with matplot lib.
Comment: Do not use any aiida methods, otherwise the methods in here can become tricky to use inside a virtual environment. Make the user extract thing out of aiida objects before hand or write something on top. Since usually parameter nodes, or files are plotted, parse a dict or filepath.
Each of the plot_methods can take keyword arguments to modify parameters of the plots There are keywords that are handled by a special class for defaults. All other arguments will be passed on to the matplotlib plotting calls
For the definition of the defaults refer to MatplotlibPlotter
-
masci_tools.vis.plot_methods.CDF_voigt_profile(x, fwhm_g, fwhm_l, mu)[source]¶ Cumulative distribution function of a voigt profile implementation of formula found here: https://en.wikipedia.org/wiki/Voigt_profile # TODO: is there an other way then to calc 2F2? # or is there an other way to calc the integral of wofz directly, or use different error functions.
-
masci_tools.vis.plot_methods.asymmetric_lorentz(x, fwhm, mu, alpha=1.0, beta=1.5)[source]¶ asymetric lorentz function
L^alpha for x<=mu L^beta for x>mu See casexps LA
-
masci_tools.vis.plot_methods.asymmetric_lorentz_gauss_conv(x, mu, fwhm_l, fwhm_g, alpha=1.0, beta=1.5)[source]¶ asymmetric Lorentzian with Gauss convoluted
-
masci_tools.vis.plot_methods.asymmetric_lorentz_gauss_sum(x, mu, fwhm_l, fwhm_g, alpha=1.0, beta=1.5)[source]¶ asymmetric Lorentzian with Gauss convoluted
-
masci_tools.vis.plot_methods.barchart(xdata, ydata, *, width=0.35, xlabel='x', ylabel='y', title='', bottom=None, saveas='barchart', axis=None, xerr=None, yerr=None, **kwargs)[source]¶ Create a standard bar chart plot (this should be flexible enough) to do all the basic bar chart plots.
- Parameters
xdata – arraylike data for the x coordinates of the bars
ydata – arraylike data for the heights of the bars
width – float determines the width of the bars
axis – Axes object where to add the plot
title – str, Title of the plot
xlabel – str, label for the x-axis
ylabel – str, label for the y-axis
saveas – str, filename for the saved plot
xerr – optional data for errorbar in x-direction
yerr – optional data for errorbar in y-direction
bottom – bottom values for the lowest end of the bars
Kwargs will be passed on to
masci_tools.vis.matplotlib_plotter.MatplotlibPlotter. If the arguments are not recognized they are passed on to the matplotlib function barTODO: grouped barchart (meaing not stacked)
-
masci_tools.vis.plot_methods.colormesh_plot(xdata, ydata, cdata, *, xlabel='', ylabel='', title='', saveas='colormesh', axis=None, **kwargs)[source]¶ Create plot with pcolormesh
- Parameters
xdata – arraylike, data for the x coordinate
ydata – arraylike, data for the y coordinate
cdata – arraylike, data for the color values with a colormap
xlabel – str, label written on the x axis
ylabel – str, label written on the y axis
title – str, title of the figure
saveas – str specifying the filename (without file format)
axis – Axes object, if given the plot will be applied to this object
Kwargs will be passed on to
masci_tools.vis.matplotlib_plotter.MatplotlibPlotter. If the arguments are not recognized they are passed on to the matplotlib function pcolormesh
-
masci_tools.vis.plot_methods.construct_corelevel_spectrum(coreleveldict, natom_typesdict, exp_references={}, scale_to=- 1, fwhm_g=0.6, fwhm_l=0.1, energy_range=[None, None], xspec=None, energy_grid=0.2, peakfunction='voigt', alpha_l=1.0, beta_l=1.5)[source]¶ Constructrs a corelevel spectrum from a given corelevel dict
- Params
- Returns
list: [xdata_spec, ydata_spec, ydata_single_all, xdata_all, ydata_all, xdatalabel]
-
masci_tools.vis.plot_methods.default_histogram(*args, **kwargs)[source]¶ Create a standard looking histogram (DEPRECATED)
-
masci_tools.vis.plot_methods.doniach_sunjic(x, scale=1.0, E_0=0, gamma=1.0, alpha=0.0)[source]¶ Doniach Sunjic asymmetric peak function. tail to higher binding energies.
param x: list values to evaluate this function param scale: multiply the function with this factor param E_0: position of the peak param gamma, ‘lifetime’ broadening param alpha: ‘asymmetry’ parametera
See Doniach S. and Sunjic M., J. Phys. 4C31, 285 (1970) or http://www.casaxps.com/help_manual/line_shapes.htm
-
masci_tools.vis.plot_methods.gauss_one(x, fwhm, mu)[source]¶ Returns a Lorentzian line shape at x with FWHM fwhm and mean mu
-
masci_tools.vis.plot_methods.gaussian(x, fwhm, mu)[source]¶ Returns Gaussian line shape at x with FWHM fwhm and mean mu
-
masci_tools.vis.plot_methods.get_mpl_help(key)[source]¶ Print the decription of the given key in the matplotlib backend
Available defaults can be seen in
MatplotlibPlotter
-
masci_tools.vis.plot_methods.histogram(xdata, density=False, histtype='bar', align='mid', orientation='vertical', log=False, axis=None, title='hist', xlabel='bins', ylabel='counts', saveas='histogram', return_hist_output=False, **kwargs)[source]¶ Create a standard looking histogram
- Parameters
xdata – arraylike, Data for the histogram
density – bool, if True the histogram is normed and a normal distribution is plotted with the same mu and sigma as the data
histtype – str, type of the histogram
align – str, defines where the bars for the bins are aligned
orientation – str, is the histogram vertical or horizontal
log – bool, if True a logarithmic scale is used for the counts
axis – Axes object where to add the plot
title – str, Title of the plot
xlabel – str, label for the x-axis
ylabel – str, label for the y-axis
saveas – str, filename for the saved plot
return_hist_output – bool, if True the data output from hist will be returned
Kwargs will be passed on to
masci_tools.vis.matplotlib_plotter.MatplotlibPlotter. If the arguments are not recognized they are passed on to the matplotlib function hist
-
masci_tools.vis.plot_methods.hyp2f2(a, b, z)[source]¶ Calculation of the 2F2() hypergeometric function, since it is not part of scipy with the identity 2. from here: https://en.wikipedia.org/wiki/Generalized_hypergeometric_function a, b,z array like inputs TODO: not clear to me how to do this… the identity is only useful if we mange the adjust the arguments in a way that we can use them… also maybe go for the special case we need first: 1,1,3/2;2;-z2
-
masci_tools.vis.plot_methods.lorentzian(x, fwhm, mu)[source]¶ Returns a Lorentzian line shape at x with FWHM fwhm and mean mu
-
masci_tools.vis.plot_methods.lorentzian_one(x, fwhm, mu)[source]¶ Returns a Lorentzian line shape at x with FWHM fwhm and mean mu
-
masci_tools.vis.plot_methods.multi_scatter_plot(xdata, ydata, *, size_data=None, color_data=None, xlabel='', ylabel='', title='', saveas='mscatterplot', axis=None, **kwargs)[source]¶ Create a scatter plot with varying marker size Info: x, y, size and color data must have the same dimensions.
- Parameters
xdata – arraylike, data for the x coordinate
ydata – arraylike, data for the y coordinate
size_data – arraylike, data for the markersizes (optional)
color_data – arraylike, data for the color values with a colormap (optional)
xlabel – str, label written on the x axis
ylabel – str, label written on the y axis
title – str, title of the figure
saveas – str specifying the filename (without file format)
axis – Axes object, if given the plot will be applied to this object
xerr – optional data for errorbar in x-direction
yerr – optional data for errorbar in y-direction
Kwargs will be passed on to
masci_tools.vis.matplotlib_plotter.MatplotlibPlotter. If the arguments are not recognized they are passed on to the matplotlib function scatter
-
masci_tools.vis.plot_methods.multiaxis_scatterplot(xdata, ydata, *, axes_loc, xlabel, ylabel, title, num_cols=1, num_rows=1, saveas='mscatterplot', **kwargs)[source]¶ Create a scatter plot with multiple axes.
- Parameters
xdata – list of arraylikes, passed on to the plotting functions for each axis (x-axis)
ydata – list of arraylikes, passed on to the plotting functions for each axis (y-axis)
axes_loc – list of tuples of two integers, location of each axis
xlabel – str or list of str, labels for the x axis
ylabel – str or list of str, labels for the y-axis
title – str or list of str, titles for the subplots
num_rows – int, how many rows of axis are created
num_cols – int, how many columns of axis are created
saveas – str filename of the saved file
- Special Kwargs:
- param subplot_params
dict with integer keys, can contain all valid kwargs for
multiple_scatterplots()with the integer key denoting to which subplot the changes are applied- param axes_kwargs
dict with integer keys, additional arguments to pass on to subplot2grid for the creation of each axis (e.g colspan, rowspan)
Other Kwargs will be passed on to all
multiple_scatterplots()calls (If they are not overwritten by parameters in subplot_params).
-
masci_tools.vis.plot_methods.multiple_scatterplots(xdata, ydata, *, xlabel='', ylabel='', title='', saveas='mscatterplot', axis=None, xerr=None, yerr=None, area_curve=None, **kwargs)[source]¶ Create a standard scatter plot with multiple sets of data (this should be flexible enough) to do all the basic plots.
- Parameters
xdata – arraylike, data for the x coordinate
ydata – arraylike, data for the y coordinate
xlabel – str, label written on the x axis
ylabel – str, label written on the y axis
title – str, title of the figure
saveas – str specifying the filename (without file format)
axis – Axes object, if given the plot will be applied to this object
xerr – optional data for errorbar in x-direction
yerr – optional data for errorbar in y-direction
area_curve – if an area plot is made this arguments defines the other enclosing line defaults to 0
Kwargs will be passed on to
masci_tools.vis.matplotlib_plotter.MatplotlibPlotter. If the arguments are not recognized they are passed on to the matplotlib functions (errorbar or fill_between)
-
masci_tools.vis.plot_methods.multiplot_moved(xdata, ydata, *, xlabel='', ylabel='', title='', scale_move=1.0, min_add=0, saveas='mscatterplot', **kwargs)[source]¶ Plots all the scatter plots above each other. It adds an arbitrary offset to the ydata to do this and calls multiple_scatterplots. Therefore you might not want to show the yaxis ticks
- Parameters
xdata – arraylike, data for the x coordinate
ydata – arraylike, data for the y coordinate
xlabel – str, label written on the x axis
ylabel – str, label written on the y axis
title – str, title of the figure
scale_move – float, max*scale_move determines size of the shift
min_add – float, minimum shift
saveas – str specifying the filename (without file format)
Kwargs are passed on to the
multiple_scatterplots()call
-
masci_tools.vis.plot_methods.plot_bands(kpath, bands, *, size_data=None, special_kpoints=None, e_fermi=0, xlabel='', ylabel='$E-E_F$ [eV]', title='', saveas='bandstructure', markersize_min=0.5, markersize_scaling=5.0, **kwargs)[source]¶ Plot the provided data for a bandstrucuture (non spin-polarized). Can be used to illustrate weights on bands via size_data
- Parameters
kpath – arraylike data for the kpoint data
bands – arraylike data for the eigenvalues
size_data – arraylike data the weights to emphasize (optional)
title – str, Title of the plot
xlabel – str, label for the x-axis
ylabel – str, label for the y-axis
saveas – str, filename for the saved plot
e_fermi – float (default 0), place the line for the fermi energy at this value
special_kpoints – list of tuples (str, float), place vertical lines at the given values and mark them on the x-axis with the given label
markersize_min – minimum value used in scaling points for weight
markersize_scaling – factor used in scaling points for weight
All other Kwargs are passed on to the
multi_scatter_plot()call
-
masci_tools.vis.plot_methods.plot_bands_and_dos()[source]¶ PLot a Bandstructure with a density of states on the right side.
-
masci_tools.vis.plot_methods.plot_certain_bands()[source]¶ Plot only certain bands from a bands.1 file from FLEUR
-
masci_tools.vis.plot_methods.plot_colortable(colors: Dict, title: str, sort_colors: bool = True, emptycols: int = 0)[source]¶ Plot a legend of named colors.
Reference: https://matplotlib.org/3.1.0/gallery/color/named_colors.html
- Parameters
colors – a dict color_name : color_value (hex str, rgb tuple, …)
title – plot title
sort_colors – sort
emptycols –
- Returns
figure
-
masci_tools.vis.plot_methods.plot_convergence_results(iteration, distance, total_energy, *, saveas1='t_energy_convergence', axis1=None, saveas2='distance_convergence', axis2=None, **kwargs)[source]¶ Plot the total energy versus the scf iteration and plot the distance of the density versus iterations.
- Parameters
iteration – array for the number of iterations
distance – array of distances
total_energy – array of total energies
saveas1 – str, filename for the energy convergence plot
axis1 – Axes object for the energy convergence plot
saveas2 – str, filename for the distance plot
axis2 – Axes object for the distance plot
Other Kwargs will be passed on to all
single_scatterplot()calls
-
masci_tools.vis.plot_methods.plot_convergence_results_m(iterations, distances, total_energies, *, modes, nodes=None, saveas1='t_energy_convergence', saveas2='distance_convergence', axis1=None, axis2=None, **kwargs)[source]¶ Plot the total energy versus the scf iteration and plot the distance of the density versus iterations.
- Parameters
iterations – array for the number of iterations
distances – array of distances
total_energies – array of total energies
modes – list of convergence modes (if ‘force’ is in the list the last distance is removed)
saveas1 – str, filename for the energy convergence plot
axis1 – Axes object for the energy convergence plot
saveas2 – str, filename for the distance plot
axis2 – Axes object for the distance plot
Other Kwargs will be passed on to all
multiple_scatterplots()calls
-
masci_tools.vis.plot_methods.plot_convex_hull2d(hull, *, title='Convex Hull', xlabel='Atomic Procentage', ylabel='Formation energy / atom [eV]', saveas='convex_hull', axis=None, **kwargs)[source]¶ Plot method for a 2d convex hull diagramm
- Parameters
hull – pyhull.Convexhull #scipy.spatial.ConvexHull
axis – Axes object where to add the plot
title – str, Title of the plot
xlabel – str, label for the x-axis
ylabel – str, label for the y-axis
saveas – str, filename for the saved plot
- Function specific parameters:
- param marker_hull
defaults to marker, marker type for the hull plot
- param markersize_hull
defaults to markersize, markersize for the hull plot
- param color_hull
defaults to color, color for the hull plot
Kwargs will be passed on to
masci_tools.vis.matplotlib_plotter.MatplotlibPlotter. If the arguments are not recognized they are passed on to the matplotlib functions plot
-
masci_tools.vis.plot_methods.plot_corelevel_spectra(coreleveldict, natom_typesdict, exp_references={}, scale_to=- 1, show_single=True, show_ref=True, energy_range=(None, None), title='', fwhm_g=0.6, fwhm_l=0.1, energy_grid=0.2, peakfunction='voigt', linestyle_spec='-', marker_spec='o', color_spec='k', color_single='g', xlabel='Binding energy [eV]', ylabel='Intensity [arb] (natoms*nelectrons)', saveas=None, xspec=None, alpha_l=1.0, beta_l=1.0, **kwargs)[source]¶ Plotting function of corelevel in the form of a spectrum.
Convention: Binding energies are positiv!
- Args:
coreleveldict: dict of corelevels with a list of corelevel energy of atomstypes # (The given corelevel accounts for a weight (number of electrons for full occupied corelevel) in the plot.) natom_typesdict: dict with number of atom types for each entry
- Kwargs:
exp_references: dict with experimental refereces, will be ploted as vertical lines show_single (bool): plot all single peaks. scale_to float: the maximum ‘intensity’ will be scaled to this value (useful for experimental comparisons) title (string): something for labeling fwhm (float): full width half maximum of peaks (gaus, lorentz or voigt_profile) energy_grid (float): energy resolution linetyp_spec : linetype for spectrum peakfunction (string): what the peakfunction should be {‘voigt’, ‘pseudo-voigt’, ‘lorentz’, ‘gaus’}
- example:
coreleveldict = {u’Be’: {‘1s1/2’ : [-1.0220669053033051, -0.3185614920138805,-0.7924091040092139]}} n_atom_types_Be12Ti = {‘Be’ : [4,4,4]}
-
masci_tools.vis.plot_methods.plot_corelevels(coreleveldict, compound='', axis=None, saveas='scatterplot', **kwargs)[source]¶ Ploting function to visualize corelevels and corelevel shifts
-
masci_tools.vis.plot_methods.plot_dos(energy_grid, dos_data, *, saveas='dos_plot', energy_label='$E-E_F$ [eV]', dos_label='DOS [1/eV]', title='Density of states', xyswitch=False, e_fermi=0, **kwargs)[source]¶ Plot the provided data for a density of states (not spin-polarized). Can be done horizontally or vertical via the switch xyswitch
- Parameters
energy_grid – arraylike data for the energy grid of the DOS
dos_data – arraylike data for all the DOS components to plot
title – str, Title of the plot
energy_label – str, label for the energy-axis
dos_label – str, label for the DOS-axis
saveas – str, filename for the saved plot
e_fermi – float (default 0), place the line for the fermi energy at this value
xyswitch – bool if True, the enrgy axis will be plotted vertically
All other Kwargs are passed on to the
multiple_scatterplots()call
-
masci_tools.vis.plot_methods.plot_lattice_constant(scaling, total_energy, *, fit_y=None, relative=True, ref_const=None, multi=False, title='Equation of states', saveas='lattice_constant', axis=None, **kwargs)[source]¶ Plot a lattice constant versus Total energy Plot also the fit. On the x axis is the scaling, it
- Parameters
scaling – arraylike, data for the scaling factor
total_energy – arraylike, data for the total energy
fit_y – arraylike, optional data of fitted data
relative – bool, scaling factor given (True), or lattice constants given?
ref_const – float (optional), or list of floats, lattice constant for scaling 1.0
multi – bool default False are they multiple plots?
- Function specific parameters:
- param marker_fit
defaults to marker, marker type for the fit data
- param markersize_fit
defaults to markersize, markersize for the fit data
- param linewidth_fit
defaults to linewidth, linewidth for the fit data
- param plotlabel_fit
str label for the fit data
Other Kwargs will be passed on to all
single_scatterplot()ormultiple_scatterplots()calls
-
masci_tools.vis.plot_methods.plot_one_element_corelv(corelevel_dict, element, compound='', axis=None, saveas='scatterplot', **kwargs)[source]¶ This routine creates a plot which visualizes all the binding energies of one element (and currenlty one corelevel) for different atomtypes.
- example:
corelevels = {‘W’ : {‘4f7/2’ : [123, 123.3, 123.4 ,123.1], ‘4f5/2’ : [103, 103.3, 103.4, 103.1]}, ‘Be’ : {‘1s’: [118, 118.2, 118.4, 118.1, 118.3]}}
-
masci_tools.vis.plot_methods.plot_relaxation_results()[source]¶ Plot from the result node of a relaxation workflow, All forces of every atom type versus relaxation cycle. Average force of all atom types versus relaxation cycle. Absolut relaxation in Angstroem of every atom type. Relative realxation of every atom type to a reference structure. (if none given use the structure from first relaxation cycle as reference)
-
masci_tools.vis.plot_methods.plot_residuen(xdata, fitdata, realdata, *, errors=None, xlabel='Energy [eV]', ylabel='cts/s [arb]', title='Residuen', saveas='residuen', hist=True, return_residuen_data=True, **kwargs)[source]¶ Calculates and plots the residuen for given xdata fit results and the real data.
If hist=True also the normed residual distribution is ploted with a normal distribution.
- Parameters
xdata – arraylike data for the x-coordinate
fitdata – arraylike fitted data for the y-coordinate
realdata – arraylike data to plot residuen against the fit
errors – dict, can be used to provide errordata for the x and y direction
xlabel – str, label for the x-axis
ylabel – str, label for the y-axis
title – str, title for the plot
saveas – str, filename for the saved plot
hist – bool, if True a normed residual distribution is ploted with a normal distribution.
return_residuen_data – bool, if True in addition to the produced axis object also the residuen data is returned
- Special Kwargs:
- param hist_kwargs
dict, these arguments will be passed on to the
histogram()call (if hist=True)
Other Kwargs will be passed on to all
single_scatterplot()call
-
masci_tools.vis.plot_methods.plot_spinpol_bands(kpath, bands_up, bands_dn, *, size_data=None, show_spin_pol=True, special_kpoints=None, e_fermi=0, xlabel='', ylabel='$E-E_F$ [eV]', title='', saveas='bandstructure', markersize_min=0.5, markersize_scaling=5.0, **kwargs)[source]¶ Plot the provided data for a bandstrucuture (spin-polarized). Can be used to illustrate weights on bands via size_data
- Parameters
kpath – arraylike data for the kpoint data
bands_up – arraylike data for the eigenvalues (spin-up)
bands_dn – arraylike data for the eigenvalues (spin-dn)
size_data – arraylike data the weights to emphasize BOTH SPINS (optional)
title – str, Title of the plot
xlabel – str, label for the x-axis
ylabel – str, label for the y-axis
saveas – str, filename for the saved plot
e_fermi – float (default 0), place the line for the fermi energy at this value
special_kpoints – list of tuples (str, float), place vertical lines at the given values and mark them on the x-axis with the given label
markersize_min – minimum value used in scaling points for weight
markersize_scaling – factor used in scaling points for weight
show_spin_pol – bool, if True (default) the two different spin channles will be shown in blue and red by default
All other Kwargs are passed on to the
multi_scatter_plot()call
-
masci_tools.vis.plot_methods.plot_spinpol_dos(energy_grid, spin_up_data, spin_dn_data, *, saveas='spinpol_dos_plot', energy_label='$E-E_F$ [eV]', dos_label='DOS [1/eV]', title='Density of states', xyswitch=False, energy_grid_dn=None, e_fermi=0, spin_dn_negative=True, **kwargs)[source]¶ Plot the provided data for a density of states (spin-polarized). Can be done horizontally or vertical via the switch xyswitch
- Parameters
energy_grid – arraylike data for the energy grid of the DOS
spin_up_data – arraylike data for all the DOS spin-up components to plot
spin_dn_data – arraylike data for all the DOS spin-down components to plot
title – str, Title of the plot
energy_label – str, label for the energy-axis
dos_label – str, label for the DOS-axis
saveas – str, filename for the saved plot
e_fermi – float (default 0), place the line for the fermi energy at this value
xyswitch – bool if True, the enrgy axis will be plotted vertically
energy_grid_dn – arraylike data for the energy grid of the DOS of the spin-down component (optional)
spin_dn_negative – bool, if True (default) the spin-down components are plotted downwards
All other Kwargs are passed on to the
multiple_scatterplots()call
-
masci_tools.vis.plot_methods.pseudo_voigt_profile(x, fwhm_g, fwhm_l, mu, mix=0.5)[source]¶ Linear combination of gaussian and loretzian instead of convolution
- Args:
x: array of floats fwhm_g: FWHM of gaussian fwhm_l: FWHM of Lorentzian mu: Mean mix: ratio of gaus to lorentz, mix* gaus, (1-mix)*Lorentz
-
masci_tools.vis.plot_methods.reset_mpl_plot_defaults()[source]¶ Reset the defaults for matplotib backend to the hardcoded defaults
Available defaults can be seen in
MatplotlibPlotter
-
masci_tools.vis.plot_methods.set_mpl_plot_defaults(**kwargs)[source]¶ Set defaults for matplotib backend according to the given keyword arguments
Available defaults can be seen in
MatplotlibPlotter
-
masci_tools.vis.plot_methods.show_mpl_plot_defaults()[source]¶ Show the currently set defaults for matplotib backend to the hardcoded defaults
Available defaults can be seen in
MatplotlibPlotter
-
masci_tools.vis.plot_methods.single_scatterplot(xdata, ydata, *, xlabel='', ylabel='', title='', saveas='scatterplot', axis=None, xerr=None, yerr=None, area_curve=None, **kwargs)[source]¶ Create a standard scatter plot (this should be flexible enough) to do all the basic plots.
- Parameters
xdata – arraylike, data for the x coordinate
ydata – arraylike, data for the y coordinate
xlabel – str, label written on the x axis
ylabel – str, label written on the y axis
title – str, title of the figure
saveas – str specifying the filename (without file format)
axis – Axes object, if given the plot will be applied to this object
xerr – optional data for errorbar in x-direction
yerr – optional data for errorbar in y-direction
area_curve – if an area plot is made this arguments defines the other enclosing line defaults to 0
Kwargs will be passed on to
masci_tools.vis.matplotlib_plotter.MatplotlibPlotter. If the arguments are not recognized they are passed on to the matplotlib functions (errorbar or fill_between)
-
masci_tools.vis.plot_methods.surface_plot(xdata, ydata, zdata, *, xlabel='', ylabel='', zlabel='', title='', saveas='surface_plot', axis=None, **kwargs)[source]¶ Create a standard surface plot
- Parameters
xdata – arraylike, data for the x coordinate
ydata – arraylike, data for the y coordinate
zdata – arraylike, data for the z coordinate
xlabel – str, label written on the x axis
ylabel – str, label written on the y axis
zlabel – str, label written on the z axis
title – str, title of the figure
axis – Axes object, if given the plot will be applied to this object
saveas – str specifying the filename (without file format)
Kwargs will be passed on to
masci_tools.vis.matplotlib_plotter.MatplotlibPlotter. If the arguments are not recognized they are passed on to the matplotlib function plot_surface
-
masci_tools.vis.plot_methods.voigt_profile(x, fwhm_g, fwhm_l, mu)[source]¶ Return the Voigt line shape at x with Lorentzian component FWHM fwhm_l and Gaussian component FWHM fwhm_g and mean mu. There is no closed form for the Voigt profile, but it is related to the real part of the Faddeeva function (wofz), which is used here.
-
masci_tools.vis.plot_methods.waterfall_plot(xdata, ydata, zdata, *, xlabel='', ylabel='', zlabel='', title='', saveas='waterfallplot', axis=None, **kwargs)[source]¶ Create a standard waterfall plot
- Parameters
xdata – arraylike, data for the x coordinate
ydata – arraylike, data for the y coordinate
zdata – arraylike, data for the z coordinate
xlabel – str, label written on the x axis
ylabel – str, label written on the y axis
zlabel – str, label written on the z axis
title – str, title of the figure
axis – Axes object, if given the plot will be applied to this object
saveas – str specifying the filename (without file format)
Kwargs will be passed on to
masci_tools.vis.matplotlib_plotter.MatplotlibPlotter. If the arguments are not recognized they are passed on to the matplotlib function scatter3D
Bokeh¶
Here the masci_tools.vis.Plotter subclass for the bokeh plotting backend
is defined with default values and many helper methods
-
class
masci_tools.vis.bokeh_plotter.BokehPlotter(**kwargs)[source]¶ Class for plotting parameters and standard code snippets for plotting with the bokeh backend.
Kwargs in the __init__ method are forwarded to setting default values for the instance
For specific documentation about the parameter/defaults handling refer to
Plotter.Below the current defined default values are shown:
Plot Parameters¶ Name
Description
Default value
figure_kwargsParameters for creating the bokeh figure. Includes things like axis type (x and y), tools, tooltips, plot width/height
{‘tools’: ‘hover’, ‘y_axis_type’: ‘linear’, ‘x_axis_type’: ‘linear’, ‘toolbar_location’: None, ‘tooltips’: [(‘X value’, ‘@x’), (‘Y value’, ‘@y’)]}
axis_linewidthLinewidth for the lines of the axis
2label_fontsizeFontsize for the labels of the axis
18pttick_label_fontsizefontsize for the ticks on the axis
16ptbackground_fill_colorColor of the background of the plot
#ffffffx_axis_formatterIf set this formatter will be used for the ticks on the x-axis
No Default
y_axis_formatterIf set this formatter will be used for the ticks on the y-axis
No Default
x_ticksTick specification for the x-axis
No Default
x_ticklabels_overwriteOverrides the labels for the ticks on the x-axis
No Default
y_ticksTick specification for the y-axis
No Default
y_ticklabels_overwriteOverrides the labels for the ticks on the y-axis
No Default
x_range_paddingSpecifies the amount of padding on the edges of the x-axis
No Default
y_range_paddingSpecifies the amount of padding on the edges of the y-axis
No Default
limitsDict specifying the limits of the axis, e.g {‘x’: (-5,5)}
No Default
legend_locationLocation of the legend inside the plot area
top_rightlegend_click_policyPolicy for what happens when labels are clicked in the legend
hidelegend_orientationOrientation of the legend
verticallegend_font_sizeFontsize for the labels inside the legend
14ptlegend_outside_plot_areaIf True the legend will be placed outside of the plot area
Falsecolor_paletteColor palette to use for the plot(s)
No Default
colorSpecific colors to use for the plot(s)
No Default
legend_labelLabels to use for the legend of the plot(s)
No Default
alphaTransparency to use for the plot(s)
1.0nameName used for identifying elements in the plot (not shown only internally)
No Default
line_colorColor to use for line plot(s)
No Default
line_alphaTransparency to use for line plot(s)
1.0line_dashDash styles to use for line plot(s)
No Default
line_widthLine width to use for line plot(s)
2.0markerType of marker to use for scatter plot(s)
circlemarker_sizeMarker size to use for scatter plot(s)
6area_plotIf True h(v)area will be used to produce the plot(s)
Falsearea_verticalDetermines, whether to use harea (False) or varea (True) for area plots
Falsefill_alphaTransparency to use for the area in area plot(s)
1.0fill_colorColor to use for the area in area plot(s)
No Default
levelCan be used to specified, which elements are fore- or background
No Default
straight_linesDict specifying straight help-lines to draw. For example {‘vertical’: 0, ‘horizontal’: [-1,1]} will draw a vertical line at 0 and two horizontal at -1 and 1
No Default
straight_line_optionsColor, width, and more options for the help-lines
{‘line_color’: ‘black’, ‘line_width’: 1.0, ‘line_dash’: ‘dashed’}
save_plotsIf True plots will be saved to file (NOT IMPLEMENTED)
FalseshowIf True bokeh.io.show will be called after the plotting routine
True-
draw_straight_lines(fig)[source]¶ Draw horizontal and vertical lines specified in the lines argument
- Parameters
fig – bokeh figure on which to perform the operation
-
plot_kwargs(ignore=None, extra_keys=None, plot_type='default', post_process=True, **kwargs)[source]¶ Creates a dict or list of dicts (for multiple plots) with the defined parameters for the plotting calls fo matplotlib
- Parameters
ignore – str or list of str (optional), defines keys to ignore in the creation of the dict
extra_keys – optional set for addtional keys to retrieve
post_process – bool, if True the parameters are cleaned up for inserting them directly into bokeh plotting functions
Kwargs are used to replace values by custom parameters:
Example for using a custom markersize:
p = BokehPlotter() p.add_parameter('marker_custom', default_from='marker') p.plot_kwargs(marker='marker_custom')
This code snippet will return the standard parameters for a plot, but the value for the marker will be taken from the key marker_custom
-
prepare_figure(title, xlabel, ylabel, figure=None)[source]¶ Create a bokeh figure according to the set parameters or modify an existing one
- Parameters
title – title of the figure
xlabel – label on the x-axis
ylabel – label on the y-axis
figure – bokeh figure, optional, if given the operations are performed on the object otherwise a new figure is created
- Returns
the created or modified bokeh figure
-
save_plot(figure)[source]¶ Show/save the bokeh figure (atm only show)
- Parameters
figure – bokeh figure on which to perform the operation
-
set_color_palette_by_num_plots()[source]¶ Set the colormap for the configured number of plots according to the set colormap or color
copied from https://github.com/PatrikHlobil/Pandas-Bokeh/blob/master/pandas_bokeh/plot.py credits to PatrikHlobil modified for use in this Plotter class
-
Here are general and special bokeh plots to use
-
masci_tools.vis.bokeh_plots.bokeh_bands(bandsdata, *, k_label='kpath', eigenvalues='eigenvalues_up', weight=None, xlabel='', ylabel='E-E_F [eV]', title='', special_kpoints=None, size_min=3.0, size_scaling=10.0, outfilename='bands_plot.html', **kwargs)[source]¶ Create an interactive bandstructure plot (non-spinpolarized) with bokeh Can make a simple plot or weight the size and color of the points against a given weight
- Parameters
bandsdata – source for the bandsdata of the plot (pandas Dataframe for example)
k_label – key from which to pull the data for the kpoints
eigenvalues – key from which to pull the data for eigenvalues
weight – optional key from the bandsdata. If given the size and color of each point are adjusted to show the weights
xlabel – label for the x-axis (default no label)
ylabel – label for the y-axis
title – title of the figure
special_kpoints – list of tuples (str, float), place vertical lines at the given values and mark them on the x-axis with the given label
e_fermi – float, determines, where to put the line for the fermi energy
size_min – minimum value used in scaling points for weight
size_scaling – factor used in scaling points for weight
outfilename – filename of the output file
Kwargs will be passed on to
bokeh_multi_scatter()
-
masci_tools.vis.bokeh_plots.bokeh_dos(dosdata, *, energy='energy_grid', ynames=None, energy_label='E-E_F [eV]', dos_label='DOS [1/eV]', title='Density of states', xyswitch=False, e_fermi=0, outfilename='dos_plot.html', **kwargs)[source]¶ Create an interactive dos plot (non-spinpolarized) with bokeh Both horizontal or vertical orientation are possible
- Parameters
dosdata – source for the dosdata of the plot (pandas Dataframe for example)
energy – key from which to pull the data for the energy grid
ynames – keys from which to pull the data for dos components
energy_label – label for the energy-axis
dos_label – label for the dos-axis
title – title of the figure
xyswitch – bool if True, the energy will be plotted along the y-direction
e_fermi – float, determines, where to put the line for the fermi energy
outfilename – filename of the output file
Kwargs will be passed on to
bokeh_line()
-
masci_tools.vis.bokeh_plots.bokeh_line(source, *, xdata='x', ydata='y', figure=None, xlabel='x', ylabel='y', title='', outfilename='scatter.html', plot_points=False, **kwargs)[source]¶ Create an interactive multi-line plot with bokeh
- Parameters
source – source for the data of the plot (pandas Dataframe for example)
xdata – key from which to pull the data for the x-axis (or if source is None list with data for x-axis)
ydata – key from which to pull the data for the y-axis (or if source is None list with data for y-axis)
xlabel – label for the x-axis
ylabel – label for the y-axis
title – title of the figure
figure – bokeh figure (optional), if provided the plot will be added to this figure
outfilename – filename of the output file
plot_points – bool, if True also plot the points with a scatterplot on top
Kwargs will be passed on to
masci_tools.vis.bokeh_plotter.BokehPlotter. If the arguments are not recognized they are passed on to the bokeh function line
-
masci_tools.vis.bokeh_plots.bokeh_multi_scatter(source, *, xdata='x', ydata='y', figure=None, xlabel='x', ylabel='y', title='', outfilename='scatter.html', **kwargs)[source]¶ Create an interactive scatter (muliple data sets possible) plot with bokeh
- Parameters
source – source for the data of the plot (pandas Dataframe for example)
xdata – key from which to pull the data for the x-axis (or if source is None list with data for x-axis)
ydata – key from which to pull the data for the y-axis (or if source is None list with data for y-axis)
xlabel – label for the x-axis
ylabel – label for the y-axis
title – title of the figure
figure – bokeh figure (optional), if provided the plot will be added to this figure
outfilename – filename of the output file
Kwargs will be passed on to
masci_tools.vis.bokeh_plotter.BokehPlotter. If the arguments are not recognized they are passed on to the bokeh function scatter
-
masci_tools.vis.bokeh_plots.bokeh_scatter(source, *, xdata='x', ydata='y', xlabel='x', ylabel='y', title='', figure=None, outfilename='scatter.html', **kwargs)[source]¶ Create an interactive scatter plot with bokeh
- Parameters
source – source for the data of the plot (pandas Dataframe for example)
xdata – key from which to pull the data for the x-axis
ydata – key from which to pull the data for the y-axis
xlabel – label for the x-axis
ylabel – label for the y-axis
title – title of the figure
figure – bokeh figure (optional), if provided the plot will be added to this figure
outfilename – filename of the output file
Kwargs will be passed on to
masci_tools.vis.bokeh_plotter.BokehPlotter. If the arguments are not recognized they are passed on to the bokeh function scatter
-
masci_tools.vis.bokeh_plots.bokeh_spinpol_bands(bandsdata, *, k_label='kpath', eigenvalues=None, weight=None, xlabel='', ylabel='E-E_F [eV]', title='', special_kpoints=None, size_min=3.0, size_scaling=10.0, outfilename='bands_plot.html', **kwargs)[source]¶ Create an interactive bandstructure plot (spinpolarized) with bokeh Can make a simple plot or weight the size and color of the points against a given weight
- Parameters
bandsdata – source for the bandsdata of the plot (pandas Dataframe for example)
k_label – key from which to pull the data for the kpoints
eigenvalues – keys from which to pull the data for eigenvalues (default [‘eigenvalues_up’,’eigenvalues_down’])
weight – optional key from the bandsdata. If given the size and color of each point are adjusted to show the weights
xlabel – label for the x-axis (default no label)
ylabel – label for the y-axis
title – title of the figure
special_kpoints – list of tuples (str, float), place vertical lines at the given values and mark them on the x-axis with the given label
e_fermi – float, determines, where to put the line for the fermi energy
size_min – minimum value used in scaling points for weight
size_scaling – factor used in scaling points for weight
outfilename – filename of the output file
Kwargs will be passed on to
bokeh_multi_scatter()
-
masci_tools.vis.bokeh_plots.bokeh_spinpol_dos(dosdata, *, spin_dn_negative=True, energy='energy_grid', ynames=None, energy_label='E-E_F [eV]', dos_label='DOS [1/eV]', title='Density of states', xyswitch=False, e_fermi=0, spin_arrows=True, outfilename='dos_plot.html', **kwargs)[source]¶ Create an interactive dos plot (spinpolarized) with bokeh Both horizontal or vertical orientation are possible
- Parameters
dosdata – source for the dosdata of the plot (pandas Dataframe for example)
energy – key from which to pull the data for the energy grid
ynames – keys from which to pull the data for dos components
spin_dn_negative – bool, if True (default), the spin down components are plotted downwards
energy_label – label for the energy-axis
dos_label – label for the dos-axis
title – title of the figure
xyswitch – bool if True, the energy will be plotted along the y-direction
e_fermi – float, determines, where to put the line for the fermi energy
spin_arrows – bool, if True (default) small arrows will be plotted on the left side of the plot indicating the spin directions (if spin_dn_negative is True)
outfilename – filename of the output file
Kwargs will be passed on to
bokeh_line()
-
masci_tools.vis.bokeh_plots.get_bokeh_help(key)[source]¶ Print the decription of the given key in the bokeh backend
Available defaults can be seen in
BokehPlotter
-
masci_tools.vis.bokeh_plots.periodic_table_plot(source, display_values=[], display_positions=[], color_value=None, tooltips=[('Name', '@name'), ('Atomic number', '@{atomic number}'), ('Atomic mass', '@{atomic mass}'), ('CPK color', '$color[hex, swatch]:CPK'), ('Electronic configuration', '@{electronic configuration}')], title='', outfilename='periodictable.html', value_color_range=[None, None], log_scale=0, color_map=None, bokeh_palette='Plasma256', toolbar_location=None, tools='hover', blank_color='#c4c4c4', blank_outsiders=[True, True], include_legend=True, copy_source=True, legend_labels=None, color_bar_title=None, show=True)[source]¶ Plot function for an interactive periodic table plot. Heat map and hover tool. source must be a panda dataframe containing, period, group,
param source: pandas dataframe containing everything param tooltips: what is shown with hover tool. values have to be in source example:
Keys of panda DF. group, period symbol and atomic number or required... Index([u'atomic number', u'symbol', u'name', u'atomic mass', u'CPK', u'electronic configuration', u'electronegativity', u'atomic radius', u'ion radius', u'van der Waals radius', u'IE-1', u'EA', u'standard state', u'bonding type', u'melting point', u'boiling point', u'density', u'metal', u'year discovered', u'group', u'period', u'rmt_mean', u'rmt_std', u'number_of_occ', u'type_color', u'c_value'], dtype='object') tooltips_def = [("Name", "@name"), ("Atomic number", "@{atomic number}"), ("Atomic mass", "@{atomic mass}"), ("CPK color", "$color[hex, swatch]:CPK"), ("Electronic configuration", "@{electronic configuration}")]
param display_values: list of strings, have to match source. Values to be displayed on the element rectangles example:[“rmt_mean”, “rmt_std”, “number_of_occ”] param display_positions: list of floats, length has to match display_values, At which y offset the display values should be displayed.
-
masci_tools.vis.bokeh_plots.plot_convergence_results(iteration, distance, total_energy, *, show=True, **kwargs)[source]¶ Plot the total energy versus the scf iteration and plot the distance of the density versus iterations. Uses bokeh_line and bokeh_scatter
- Parameters
iteration – list of Int
distance – list of floats
show – bool, if True call show
- Total_energy
list of floats
Kwargs will be passed on to
bokeh_line()- Returns grid
bokeh grid with figures
-
masci_tools.vis.bokeh_plots.plot_convergence_results_m(iterations, distances, total_energies, *, link=False, nodes=None, modes=None, plot_label=None, saveas1='t_energy_convergence', saveas2='distance_convergence', show=True, **kwargs)[source]¶ Plot the total energy versus the scf iteration and plot the distance of the density versus iterations in a bokeh grid for several SCF results.
- Parameters
distances – list of lists of floats
iterations – list of lists of Int
link – bool, optional default=False:
nodes – list of node uuids or pks important for links
saveas1 – str, optional default=’t_energy_convergence’, save first figure as
saveas2 – str, optional default=’distance_convergence’, save second figure as
figure_kwargs – dict, optional default={‘plot_width’: 600, ‘plot_height’: 450}, gets parsed to bokeh_line
kwargs – further key-word arguments for bokeh_line
- Total_energies
list of lists of floats
- Returns grid
bokeh grid with figures
-
masci_tools.vis.bokeh_plots.plot_convex_hull2d(hull, title='Convex Hull', xlabel='Atomic Procentage', ylabel='Formation energy / atom [eV]', linestyle='-', marker='o', legend=True, legend_option={}, saveas='convex_hull', limits=[None, None], scale=[None, None], axis=None, color='k', color_line='k', linewidth=2, markersize=8, marker_hull='o', markersize_hull=8, **kwargs)[source]¶ Plot method for a 2d convex hull diagram
- Parameters
hull – scipy.spatial.ConvexHull
-
masci_tools.vis.bokeh_plots.reset_bokeh_plot_defaults()[source]¶ Reset the defaults for bokeh backend to the hardcoded defaults
Available defaults can be seen in
BokehPlotter
-
masci_tools.vis.bokeh_plots.set_bokeh_plot_defaults(**kwargs)[source]¶ Set defaults for bokeh backend according to the given keyword arguments
Available defaults can be seen in
BokehPlotter
-
masci_tools.vis.bokeh_plots.show_bokeh_plot_defaults()[source]¶ Show the currently set defaults for bokeh backend
Available defaults can be seen in
BokehPlotter
Calculation tools¶
This file contains a class to compute the crystalfield coefficients from convoluting the charge density with the potential which produces the crystalfield. This is both compatible with the Yttrium-Analogue approximation and self-consitent calculation of the potential
-
class
masci_tools.tools.cf_calculation.CFCalculation(radial_points=4000, reference_radius='pot', pot_cutoff=0.001, only_m0=False, quiet=False)[source]¶ Class for calculating Crystal Field coefficients using the procedure described in C.E. Patrick, J.B. Staunton: J. Phys.: Condens. Matter 31, 305901 (2019)
- Using the formula:
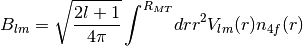
The read in quantities are interpolated from logarithmic meshes to equidistant meshes
The function constructs an equidistant mesh between 0 and the muffin tin radius defined in self.reference_radius and with self.radial_points points
- Parameters:
- param radial_points
int, number of radial points in the interpolated mesh
- param reference_radius
stror float; Either ‘pot’ or ‘cdn’ or explicit number. Defines which muffin-tin radius is used for the equidistant mesh. IMPORTANT! If txt files are used the muffin-tin radius has to be provided explicitly
- param pot_cutoff
float Defines minimum value that has to appear in potentials to not be omitted (Only HDF)
- param only_m0
bool, Ignores coefficients with m!=0 if True
- param quiet
bool, supresses print statements if True
-
performIntegration(convert=True)[source]¶ Performs the integration to obtain the crystal field coefficients If the data was not already interpolated, the interpolation will be performed beforehand
- Parameters:
- param convert
bool, converts to Steven’s coefficients (if True)
- Returns
list of CFCoefficient objects (namedtuple), with all the necessary information
-
prefactor(l, m)[source]¶ Gives the lm dependent prefactor for conversion between Blm and Alm coefficients
- Args:
- param l
int; orbital quantum number
- param m
int; magnetic quantum number
- Returns
float prefactor for conversion to Steven’s Coefficients
-
readCDN(file, **kwargs)[source]¶ Reads in the normed charge density for the CF coefficient calculation If hdf files are given also the muffin tin radius is read in
- Parameters:
- param file
Expects string filename for the charge density to read in The function expects either HDF files or txt files with the format (rmesh,cdn). The charge density should be given as r^2n(r) and normed to 1
- kwargs:
- param atomType
int, Defines the atomType to read in (only for HDF files)
- param header
int, Define how many lines to skip in the beginning of txt file
-
readPot(*args, lm=None, **kwargs)[source]¶ Reads in the potentials for the CF coefficient calculation If hdf files are given also the muffin tin radius is read in
- Parameters:
- param args
Expects string filenames for the potentials to read in The function expects either HDF files or txt files with the format (rmesh,vlmup,vlmdn)
- param lm
list of tuples, Defines the l and m indices for the given txt files
- kwargs:
- param atomType
int, Defines the atomType to read in (only for HDF files)
- param header
int, Define how many lines to skip in the beginning of txt file
- param complexData
bool, Define if the data in the text file is complex
- Raises:
ValueError: lm indices list length has to match number of files read in
-
class
masci_tools.tools.cf_calculation.CFCoefficient(l, m, spin_up, spin_down, unit, convention)¶ -
property
convention¶ Alias for field number 5
-
property
l¶ Alias for field number 0
-
property
m¶ Alias for field number 1
-
property
spin_down¶ Alias for field number 3
-
property
spin_up¶ Alias for field number 2
-
property
unit¶ Alias for field number 4
-
property
-
masci_tools.tools.cf_calculation.plot_crystal_field_calculation(cfcalc, filename='crystal_field_calc', pot_title='Potential', cdn_title='Density', xlabel='$R$ (Bohr)', pot_ylabel='$Vpot$ (Hartree)', cdn_ylabel='Density', fontsize=12, labelsize=12, pot_colors=None, save=False, show=True)[source]¶ Plot the given potentials and charge densities
- Parameters:
- param cfcalc
CFcalculation containing the data to plot
- param filename
str, Define the filename to save the figure
- param pot_title
Title for the potential subplot
- param cdn_title
Title for the charge density subplot
- param xlabel
label for the x axis of both subplots
- param pot_ylabel
label for the y axis of the potential subplot
- param cdn_ylabel
label for the y axis f the charge density subplot
- param fontsize
fontsize for titles and labels on the axis
- param labelsize
fontsize for the ticks on the axis,
-
masci_tools.tools.cf_calculation.plot_crystal_field_potential(cfcoeffs, filename='crystal_field_potential_areaplot', spin='avg', phi=0.0, save=False, show=True)[source]¶ Plots the angular dependence of the calculated CF potential as well as a plane defined by phi.
- Parameters:
- param cfcoeffs
list of CFCoefficients to construct the potential
- param filename
str, defines the filename to save the figure
- param spin
str; Either ‘up’, ‘dn’ or ‘avg’. Which spin direction to plot (‘avg’-> (‘up’+’dn’)/2.0)
- param phi
float, defines the phi angle of the plane
- Raises
AssertionError – When coefficients are provided as wrong types or in the wrong convention
IO helper functions and file parsers¶
KKR related IO¶
-
class
masci_tools.io.kkr_params.kkrparams(**kwargs)[source]¶ Class for creating and handling the parameter input for a KKR calculation Optional keyword arguments are passed to init and stored in values dictionary.
Example usage: params = kkrparams(LMAX=3, BRAVAIS=array([[1,0,0], [0,1,0], [0,0,1]]))
- Alternatively values can be set afterwards either individually with
params.set_value(‘LMAX’, 3)
- or multiple keys at once with
params.set_multiple_values(EMIN=-0.5, EMAX=1)
Other useful functions
print the description of a keyword: params.get_description([key]) where [key] is a string for a keyword in params.values
print a list of mandatory keywords: params.get_all_mandatory()
print a list of keywords that are set including their value: params.get_set_values()
-
change_XC_val_kkrimp(val)[source]¶ Convert integer value of KKRhost KEXCOR input to KKRimp XC string input.
-
fill_keywords_to_inputfile(is_voro_calc=False, output='inputcard')[source]¶ Fill new inputcard with keywords/values automatically check for input consistency if is_voro_calc==True change mandatory list to match voronoi code, default is KKRcode
-
classmethod
get_KKRcalc_parameter_defaults(silent=False)[source]¶ set defaults (defined in header of this file) and returns dict, kkrparams_version
-
get_description(key=None, search=None)[source]¶ Returns description of keyword ‘key’ If ‘key’ is None, print all descriptions of all available keywords If ‘search’ is not None, print all keys+descriptions where the search string is found
-
get_dict(group=None, subgroup=None)[source]¶ Returns values dictionary.
Prints values belonging to a certain group only if the ‘group’ argument is one of the following: ‘lattice’, ‘chemistry’, ‘accuracy’, ‘external fields’, ‘scf cycle’, ‘other’
Additionally the subgroups argument allows to print only a subset of all keys in a certain group. The following subgroups are available.
in ‘lattice’ group ‘2D mode’, ‘shape functions’
in ‘chemistry’ group ‘Atom types’, ‘Exchange-correlation’, ‘CPA mode’, ‘2D mode’
in ‘accuracy’ group ‘Valence energy contour’, ‘Semicore energy contour’, ‘CPA mode’, ‘Screening clusters’, ‘Radial solver’, ‘Ewald summation’, ‘LLoyd’
-
read_keywords_from_inputcard(inputcard='inputcard')[source]¶ Read list of keywords from inputcard and extract values to keywords dict
- Example usage
p = kkrparams(); p.read_keywords_from_inputcard(‘inputcard’)
- Note
converts ‘<RBLEFT>’, ‘<RBRIGHT>’, ‘ZPERIODL’, and ‘ZPERIODR’ automatically to Ang. units!
-
set_multiple_values(**kwargs)[source]¶ Set multiple values (in example value1 and value2 of keywords ‘key1’ and ‘key2’) given as key1=value1, key2=value2
-
classmethod
split_kkr_options(valtxt)[source]¶ Split keywords after fixed length of 8 :param valtxt: list of strings or single string :returns: List of keywords of maximal length 8
-
masci_tools.io.kkr_read_shapefun_info.read_shapefun(path='.')[source]¶ Read vertices of shapefunctions with Zoom into shapefun of a single atom
- Author
Philipp Ruessmann
- Parameters
path – path where voronoi output is found (optional, defaults to ‘./’)
- Returns pos
positions of the centers of the shapefunctions
- Returns out
dictionary of the vertices of the shapefunctions
Here I collect all functions needed to parse the output of a KKR calculation. These functions do not need aiida and are therefore separated from the actual parser file where parse_kkr_outputfile is called
-
masci_tools.io.parsers.kkrparser_functions.check_error_category(err_cat, err_msg, out_dict)[source]¶ Check if parser error of the non-critical category (err_cat != 1) are actually consistent and may be discarded.
- Parameters
err_cat – the error-category of the error message to be investigated
err_msg – the error-message
out_dict – the dict of results obtained from the parser function
- Returns
True/False if message is an error or warning
-
masci_tools.io.parsers.kkrparser_functions.get_kmeshinfo(outfile_0init, outfile_000)[source]¶ Extract kmesh info from output.0.txt and output.000.txt
-
masci_tools.io.parsers.kkrparser_functions.get_lattice_vectors(outfile_0init)[source]¶ read direct and reciprocal lattice vectors in internal units (useful for qdos generation)
-
masci_tools.io.parsers.kkrparser_functions.get_natom(outfile_0init)[source]¶ extract NATYP value from output.0.txt
-
masci_tools.io.parsers.kkrparser_functions.get_noco_rms(outfile, debug=False)[source]¶ Get average noco rms error
-
masci_tools.io.parsers.kkrparser_functions.get_nspin(outfile_0init)[source]¶ extract NSPIN value from output.0.txt
-
masci_tools.io.parsers.kkrparser_functions.get_orbmom(outfile, natom)[source]¶ read orbmom info from outfile and return array (iteration, atom)=orbmom
-
masci_tools.io.parsers.kkrparser_functions.get_rms(outfile, outfile2, debug=False)[source]¶ Get rms error per atom (both values for charge and spin) and total (i.e. average) value
-
masci_tools.io.parsers.kkrparser_functions.get_single_particle_energies(outfile_000)[source]¶ extracts single particle energies from outfile_000 (output.000.txt) returns the valence contribution of the single particle energies
-
masci_tools.io.parsers.kkrparser_functions.get_spinmom_per_atom(outfile, natom, nonco_out_file=None)[source]¶ Extract spin moment information from outfile and nonco_angles_out (if given)
-
masci_tools.io.parsers.kkrparser_functions.parse_array_float(outfile, searchstring, splitinfo, replacepair=None, debug=False)[source]¶ Search for keyword searchstring in outfile and extract array of results
Returns: array of results
-
masci_tools.io.parsers.kkrparser_functions.parse_kkr_outputfile(out_dict, outfile, outfile_0init, outfile_000, timing_file, potfile_out, nonco_out_file, outfile_2='output.2.txt', skip_readin=False, debug=False)[source]¶ Parser method for the kkr outfile. It returns a dictionary with results
-
masci_tools.io.parsers.kkrparser_functions.use_newsosol(outfile_0init)[source]¶ extract NEWSOSOL info from output.0.txt
Everything that is needed to parse the output of a voronoi calculation.
-
masci_tools.io.parsers.voroparser_functions.check_voronoi_output(potfile, outfile, delta_emin_safety=0.1)[source]¶ Read output from voronoi code and create guess of energy contour
-
masci_tools.io.parsers.voroparser_functions.get_valence_min(outfile='out_voronoi')[source]¶ Construct minimum of energy contour (between valence band bottom and core states)
-
masci_tools.io.parsers.voroparser_functions.parse_voronoi_output(out_dict, outfile, potfile, atominfo, radii, inputfile, debug=False)[source]¶ Parse output of voronoi calculation and return (success, error_messages_list, out_dict)
Tools for the impurity caluclation plugin and its workflows
-
class
masci_tools.io.parsers.kkrimp_parser_functions.KkrimpParserFunctions[source]¶ Class of parser functions for KKRimp calculation
- Usage
success, msg_list, out_dict = parse_kkrimp_outputfile().parse_kkrimp_outputfile(out_dict, files)
-
parse_kkrimp_outputfile(out_dict, file_dict, debug=False)[source]¶ Main parser function for kkrimp, read information from files in file_dict and fills out_dict :param out_dict: dictionary that is filled with parsed output of the KKRimp calculation :param file_dict: dictionary of files that are parsed :returns: success (bool), msg_list(list of error/warning messages of parser), out_dict (filled dict of parsed output) :note: file_dict should contain the following keys
‘outfile’, the std_out of the KKRimp calculation
‘out_log’, the out_log.000.txt file
‘out_pot’, the output potential
‘out_enersp_at’, the out_energysp_per_atom_eV file
‘out_enertot_at’, the out_energytotal_per_atom_eV file
‘out_timing’, the timing file
‘kkrflex_llyfac’, the file for the Lloyd factor
‘kkrflex_angles’, the nonco_angles file for the KKRimp calculation
‘out_spinmoms’, the output spin moments file
‘out_orbmoms’, the output orbital moments file
General HDF5 parser¶
This module contains a generic HDF5 reader
-
class
masci_tools.io.parsers.hdf5.reader.AttribTransformation(name, attrib_name, args, kwargs)¶ -
property
args¶ Alias for field number 2
-
property
attrib_name¶ Alias for field number 1
-
property
kwargs¶ Alias for field number 3
-
property
name¶ Alias for field number 0
-
property
-
class
masci_tools.io.parsers.hdf5.reader.HDF5Reader(file, move_to_memory=True)[source]¶ Class for reading in data from hdf5 files using a specified recipe
- Parameters
file – filepath to hdf file or opened file handle (mode ‘rb’)
move_to_memory – bool if True after reading and transforming the data all leftover h5py.Datasets are moved into np.arrays
The recipe is passed to the
HDF5Reader.read()method and consists of a dict specifiying which attributes and datasets to read in and how to transform themEach attribute/dataset entry corresponds to one entry point in the given .hdf file Available transformations can either be found in
transformsor can be defined by the user with thehdf5_transformation()decoratorBasic Usage:
from masci_tools.io.parsers.hdf5 import HDF5Reader import masci_tools.io.parsers.hdf5.recipes as recipes #This example shows the usage for producing data from a bandstructure calculation #in Fleur with HDF5Reader('/path/to/hdf/banddos.hdf') as h5reader: data, attributes = h5reader.read(recipe=recipes.FleurBands) print(data, attributes)
-
class
masci_tools.io.parsers.hdf5.reader.Transformation(name, args, kwargs)¶ -
property
args¶ Alias for field number 1
-
property
kwargs¶ Alias for field number 2
-
property
name¶ Alias for field number 0
-
property
This module defines commonly used recipes for the HDF5Reader
- Available are:
Recipe for bandstructure calculations with Fleur
Recipes for almost all DOS calculation modes of Fleur
A Recipe is a python dictionary in a specific format.
A Template Example:
from masci_tools.io.parser.hdf5.readers import Transformation, AttribTransformation
RecipeExample = {
'datasets': {
'example_dataset': {
'h5path': '/path/in/hdf/file',
'transforms': [Transformation(name='get_first_element')]
},
'example_attrib_transform': {
'h5path': '/other/path/in/hdf/file',
'transforms': [AttribTransformation(name='multiply_by_attribute', attrib_name='example_attribute')]
}
},
'attributes': {
'example_attribute': {
'h5path':
'/path/in/hdf/file',
'transforms':
[Transformation(name='get_attribute', args=('attribName',)),
Transformation(name='get_first_element')]
}
}
}
The Recipe consists of two sections ‘datasets’ and ‘attributes’. All data from these two sections will be returned
in separate python dictionaries by the HDF5Reader class
Each entry in those sections has to have a h5path entry, which will specify the dataset to initially
read from the hdf file. Then each entry can define a entry transforms with a list of the namedtuples
imported at the top of the code example. These correponds to function calls to functions in
transforms to transform the read in data
Entries in the attributes section are read and transformed first and can subsequently be used in transformations
for the datasets. These correpsond to the transforms created with the AttribTransformation
namedtuple instead of Transformation.
-
masci_tools.io.parsers.hdf5.recipes.get_fleur_bands_specific_weights(weight_name)[source]¶ Recipe for bandstructure calculations only retrieving one additional weight besides the eigenvalues and kpath
- Parameters
weight_name – key or list of keys of the weight(s) to retrieve
- Returns
dict of the recipe to retrieve a simple bandstructure plus the one specified weight
Collection of predefined transformations for the HDF5Reader class
All Transformation have to be able to handle (or fail gracefully with a clear error) for the following 3 cases:
The dataset is still a h5py.Dataset and might need to be transformed to a numpy array
The dataset is a numpy array
The dataset is a dict. This is needed to read arbitrary child dataset, where not all labels are known. Two options can be chosen apply the transformation to all keys in the dict or throw an error
-
masci_tools.io.parsers.hdf5.transforms.add_partial_sums(dataset, attribute_value, pattern_format, make_set=False)[source]¶ Add entries to the dataset dict (Only avalaible for dict datasets) with sums over entries containing a given pattern formatted with a attribute_value
Used for example in the FleurBands recipe to calculate total atom weights with the pattern_format ‘MT:{}’.format and the atomtype as the attribute_value
- Parameters
dataset – dataset to transform
attribute_value – value to multiply by (attribute value passed in from _transform_dataset)
pattern_format – callable returning a formatted string This will be called with every entry in the attribute_value list
- Returns
dataset with new entries containing the sums over entries matching the given pattern
-
masci_tools.io.parsers.hdf5.transforms.attributes(dataset)[source]¶ Extracts all attributes of the dataset
- Parameters
dataset – dataset to transform
- Returns
dict with all the set attributes on the dataset
-
masci_tools.io.parsers.hdf5.transforms.calculate_norm(dataset, between_neighbours=False)[source]¶ Calculate norms on the given dataset. Calculates the norm of each row in the dataset
- Parameters
dataset – dataset to transform
between_neighbours – bool, if True the distance between subsequent entries in the dataset is calculated
- Returns
norms of the given dataset
-
masci_tools.io.parsers.hdf5.transforms.convert_to_complex_array(dataset)[source]¶ Converts the given dataset of real numbers into dataset of complex numbers. This follows the convention of how complex numbers are normally written out by Fleur (last index 0 real part, last index 1 imag part)
- Parameters
dataset – dataset to transform
- Returns
dataset with complex values
-
masci_tools.io.parsers.hdf5.transforms.convert_to_str(dataset)[source]¶ Converts the given dataset to a numpy array of type string
- Parameters
dataset – dataset to transform
- Returns
numpy array of dtype str
-
masci_tools.io.parsers.hdf5.transforms.cumulative_sum(dataset, beginning_zero=True)[source]¶ Calculate the cumulative sum of the dataset
- Parameters
dataset – dataset to transform
- Returns
cumulative sum of the dataset
-
masci_tools.io.parsers.hdf5.transforms.flatten_array(dataset, order='C')[source]¶ Flattens the given dataset to one dimensional array. Copies the array !!
- Parameters
dataset – dataset to transform
order – str {‘C’, ‘F’, ‘A’, ‘K’} flatten in column major or row-major order (see numpy.flatten documentation)
- Returns
flattened dataset
-
masci_tools.io.parsers.hdf5.transforms.get_all_child_datasets(group, ignore=None, contains=None)[source]¶ Get all datasets contained in the given group
- Parameters
group – h5py object to extract from
ignore – str or iterable of str (optional). These keys will be ignored
- Returns
a dict with the contained dataset entered with their names as keys
-
masci_tools.io.parsers.hdf5.transforms.get_attribute(dataset, attribute_name)[source]¶ Extracts a specified attribute’s value.
- Parameters
dataset – dataset to transform
attribute_name – str of the attribute to extract from the dataset
- Returns
value of the attribute on the dataset
-
masci_tools.io.parsers.hdf5.transforms.get_first_element(dataset)[source]¶ Get the first element of the dataset.
- Parameters
dataset – dataset to transform
- Returns
first element of the dataset
-
masci_tools.io.parsers.hdf5.transforms.get_name(dataset, full_path=False)[source]¶ Get the name of the dataset.
- Parameters
dataset – dataset to get the shape
full_path – bool, if True the full path to the dataset is returned
- Returns
name of the dataset
-
masci_tools.io.parsers.hdf5.transforms.get_shape(dataset)[source]¶ Get the shape of the dataset.
- Parameters
dataset – dataset to get the shape
- Returns
shape of the dataset
-
masci_tools.io.parsers.hdf5.transforms.hdf5_transformation(*, attribute_needed)[source]¶ Decorator for registering a function as a transformation functions on the
HDF5Readerclass- Parameters
attribute_needed – bool if True this function takes a previously processed attribute value and is therefore only available for the entries in datasets
-
masci_tools.io.parsers.hdf5.transforms.index_dataset(dataset, index)[source]¶ Get the n-th element of the dataset.
- Parameters
dataset – dataset to transform
- Returns
first element of the dataset
-
masci_tools.io.parsers.hdf5.transforms.move_to_memory(dataset)[source]¶ Moves the given dataset to memory, if it’s not already there Creates numpy arrays for each dataset it finds
- Parameters
dataset – dataset to transform
- Returns
dataset with h5py.Datasets converted to numpy arrays
-
masci_tools.io.parsers.hdf5.transforms.multiply_array(dataset, matrix, transpose=False)[source]¶ Multiply the given dataset with a matrix
- Parameters
dataset – dataset to multiply
matrix – matrix to multiply by
transpose – bool, if True the given matrix is transposed
- Returns
dataset multiplied with the given matrix
-
masci_tools.io.parsers.hdf5.transforms.multiply_by_attribute(dataset, attribute_value, transpose=False)[source]¶ Multiply the given dataset with a previously parsed attribute, either scalar or matrix like
- Parameters
dataset – dataset to transform
attribute_value – value to multiply by (attribute value passed in from _transform_dataset)
- Only relevant for matrix multiplication:
- param transpose
bool if True the Matrix order is transposed before multiplying
- Returns
dataset multiplied with the given attribute_value
-
masci_tools.io.parsers.hdf5.transforms.multiply_scalar(dataset, scalar_value)[source]¶ Multiply the given dataset with a scalar_value
- Parameters
dataset – dataset to transform
scalar_value – value to mutiply the dataset by
- Returns
the dataset multiplied by the scalar if it is a dict all entries are multiplied
-
masci_tools.io.parsers.hdf5.transforms.periodic_elements(dataset)[source]¶ - Converts the given dataset (int or list of ints)
To the atomic symbols corresponding to the atomic number
- Parameters
dataset – dataset to transform
- Returns
str or array of str with the atomic elements
-
masci_tools.io.parsers.hdf5.transforms.repeat_array(dataset, n_repeats)[source]¶ Use numpy.repeat to repeat each element in array n-times
- Parameters
dataset – dataset to transform
n_repeats – int, time to repeat each element
- Returns
dataset with elements repeated n-times
-
masci_tools.io.parsers.hdf5.transforms.repeat_array_by_attribute(dataset, attribute_value)[source]¶ Use numpy.repeat to repeat each element in array n-times (given by attribute_value)
- Parameters
dataset – dataset to transform
attribute_shape – int, time to repeat the elements in the given array
- Returns
dataset with elements repeated n-times
-
masci_tools.io.parsers.hdf5.transforms.shift_by_attribute(dataset, attribute_value, negative=False)[source]¶ Shift the dataset by the given value of the attribute
- Parameters
dataset – dataset to transform
attribute_value – value to shift the dataset by
negative – bool, if True the scalar_value will be substracted
- Returns
the dataset shifted by the scalar if it is a dict all entries are shifted
-
masci_tools.io.parsers.hdf5.transforms.shift_dataset(dataset, scalar_value, negative=False)[source]¶ Shift the dataset by the given scalar_value
- Parameters
dataset – dataset to transform
scalar_value – value to shift the dataset by
negative – bool, if True the scalar_value will be substracted
- Returns
the dataset shifted by the scalar if it is a dict all entries are shifted
-
masci_tools.io.parsers.hdf5.transforms.slice_dataset(dataset, slice_arg)[source]¶ Slice the dataset with the given slice argument.
- Parameters
dataset – dataset to transform
slice_arg – slice to apply to the dataset
- Returns
first element of the dataset
-
masci_tools.io.parsers.hdf5.transforms.split_array(dataset, suffixes=None, name=None)[source]¶ Split the arrays in a dataset into multiple entries by their first index
If the dataset is a dict the entries will be split up. If the dataset is not a dict a dict is created with the dataset entered under name and this will be split up
- Parameters
dataset – dataset to transform
suffix – Optional list of str to use for suffixes for the split up entries. by default it is the value of the first index of the original array
name – str for the case of the dataset not being a dict. Key for the entry in the new dict for the original dataset. The returned dataset will only contain the split up entries
dataset – dict with the entries split up
-
masci_tools.io.parsers.hdf5.transforms.sum_over_dict_entries(dataset, overwrite_dict=False)[source]¶ Sum the datasets contained in the given dict dataset
- Parameters
dataset – dataset to transform
overwrite_dict – bool if True, the result will overwrite the dictionary if False it is entered under sum in the dict
- Returns
dataset with summed entries
Definition of default parsing tasks for fleur out.xml¶
This module contains the dictionary with all defined tasks for the outxml_parser. The entries in the TASK_DEFINITION dict specify how to parse specific attributes tags.
This needs to be maintained if the specifications do not work for a new schema version because of changed attribute names for example.
Each entry in the TASK_DEFINITION dict can contain a series of keys, which by default correspond to the keys in the output dictionary
- The following keys are expected in each entry:
- param parse_type
str, defines which methods to use when extracting the information
- param path_spec
dict with all the arguments that should be passed to get_tag_xpath or get_attrib_xpath to get the correct path
- param subdict
str, if present the parsed values are put into this key in the output dictionary
- param overwrite_last
bool, if True no list is inserted and each entry overwrites the last
- For the allAttribs parse_type there are more keys that can appear:
- param base_value
str, optional. If given the attribute with this name will be inserted into the key from the task_definition all other keys are formatted as {task_key}_{attribute_name}
- param ignore
list of str, these attributes will be ignored
- param overwrite
list of str, these attributes will not create a list and overwrite any value that might be there
- param flat
bool, if False the dict parsed from the tag is inserted as a dict into the correspondin key if True the values will be extracted and put into the output dictionary with the format {task_key}_{attribute_name}
Each task entry can have additional keys to specify, when to perform the task. These are denoted with underscores in their names and are all optional:
- param _general
bool, default False. If True the parsing is not performed for each iteration on the iteration node but beforehand and on the root node
- param _modes
list of tuples, sets conditions for the keys in fleur_modes to perform the task .e.g. [(‘jspins’, 2), (‘soc’, True)] means only perform this task for a magnetic soc calculation
- param _minimal
bool, default False, denotes task to perform when minimal_mode=True is passed to the parser
- param _special
bool, default False, If true these tasks are not added by default and need to be added manually
- param _conversions
list of str, gives the names of functions in fleur_outxml_conversions to perform after parsing
- The following keys are special at the moment:
`fleur_modes`specifies how to identify the type of the calculation (e.g. SOC, magnetic, lda+u) this is used to determine, whether additional things should be parsed
Following is the current specification of tasks
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170 171 172 173 174 175 176 177 178 179 180 181 182 183 184 185 186 187 188 189 190 191 192 193 194 195 196 197 198 199 200 201 202 203 204 205 206 207 208 209 210 211 212 213 214 215 216 217 218 219 220 221 222 223 224 225 226 227 228 229 230 231 232 233 234 235 236 237 238 239 240 241 242 243 244 245 246 247 248 249 250 251 252 253 254 255 256 257 258 259 260 261 262 263 264 265 266 267 268 269 270 271 272 273 274 275 276 277 278 279 280 281 282 283 284 285 286 287 288 289 290 291 292 293 294 295 296 297 298 299 300 301 302 303 304 305 306 307 308 309 310 311 312 313 314 315 316 317 318 319 320 321 322 323 324 325 326 327 328 329 330 331 332 333 334 335 336 337 338 339 340 341 342 343 344 345 346 347 348 349 350 351 352 353 354 355 356 357 358 359 360 361 362 363 364 365 366 367 368 369 370 371 372 373 374 375 376 377 378 379 380 381 382 383 384 385 386 387 388 389 390 391 392 393 394 395 396 397 398 399 400 401 402 403 404 405 406 407 408 409 410 411 412 413 414 415 416 417 418 419 420 421 422 423 424 425 426 427 428 429 430 431 432 433 434 435 436 437 438 439 440 441 442 443 444 445 446 447 448 449 450 451 452 453 454 455 456 457 458 459 460 461 462 463 464 465 466 467 468 469 470 471 472 473 474 475 476 477 478 479 480 481 482 483 484 485 486 487 488 489 490 491 492 493 494 495 496 497 498 499 500 501 502 503 504 505 506 507 508 509 510 511 512 513 514 515 516 517 518 519 520 521 522 523 524 525 526 527 528 529 530 531 532 533 | __working_out_versions__ = {'0.34'}
__base_version__ = '0.34'
TASKS_DEFINITION = {
#--------Definitions for general info from outfile (start, endtime, number_iterations)--------
'general_out_info': {
'_general': True,
'_minimal': True,
'_conversions': ['calculate_walltime'],
'creator_name': {
'parse_type': 'attrib',
'path_spec': {
'name': 'version',
'not_contains': 'git'
}
},
'creator_target_architecture': {
'parse_type': 'text',
'path_spec': {
'name': 'targetComputerArchitectures'
}
},
'output_file_version': {
'parse_type': 'attrib',
'path_spec': {
'name': 'fleurOutputVersion'
}
},
'number_of_iterations': {
'parse_type': 'numberNodes',
'path_spec': {
'name': 'iteration'
}
},
'number_of_atoms': {
'parse_type': 'attrib',
'path_spec': {
'name': 'nat'
}
},
'number_of_atom_types': {
'parse_type': 'attrib',
'path_spec': {
'name': 'ntype'
}
},
'number_of_kpoints': {
'parse_type': 'attrib',
'path_spec': {
'name': 'count',
'contains': 'numericalParameters'
}
},
'start_date': {
'parse_type': 'allAttribs',
'path_spec': {
'name': 'startDateAndTime'
},
'ignore': ['zone'],
'flat': False,
},
'end_date': {
'parse_type': 'allAttribs',
'path_spec': {
'name': 'endDateAndTime'
},
'ignore': ['zone'],
'flat': False,
}
},
#--------Defintions for general info from input section of outfile (kmax, symmetries, ..)--------
'general_inp_info': {
'_general': True,
'_minimal': True,
'title': {
'parse_type': 'text',
'path_spec': {
'name': 'comment'
}
},
'kmax': {
'parse_type': 'attrib',
'path_spec': {
'name': 'Kmax'
}
},
'gmax': {
'parse_type': 'attrib',
'path_spec': {
'name': 'Gmax'
}
},
'number_of_spin_components': {
'parse_type': 'attrib',
'path_spec': {
'name': 'jspins'
}
},
'number_of_symmetries': {
'parse_type': 'numberNodes',
'path_spec': {
'name': 'symOp'
}
},
'number_of_species': {
'parse_type': 'numberNodes',
'path_spec': {
'name': 'species'
}
},
'film': {
'parse_type': 'exists',
'path_spec': {
'name': 'filmPos'
}
},
},
#--------Defintions for lda+u info from input section (species, ldau tags)--------
'ldau_info': {
'_general': True,
'_modes': [('ldau', True)],
'_conversions': ['convert_ldau_definitions'],
'parsed_ldau': {
'parse_type': 'allAttribs',
'path_spec': {
'name': 'ldaU',
'contains': 'species'
},
'subdict': 'ldau_info',
'flat': False,
'only_required': True
},
'ldau_species': {
'parse_type': 'parentAttribs',
'path_spec': {
'name': 'ldaU',
'contains': 'species'
},
'subdict': 'ldau_info',
'flat': False,
'only_required': True
}
},
#--------Defintions for relaxation info from input section (bravais matrix, atompos)
#--------for Bulk and film
'bulk_relax_info': {
'_general': True,
'_modes': [('relax', True), ('film', False)],
'_conversions': ['convert_relax_info'],
'lat_row1': {
'parse_type': 'text',
'path_spec': {
'name': 'row-1',
'contains': 'bulkLattice/bravais'
}
},
'lat_row2': {
'parse_type': 'text',
'path_spec': {
'name': 'row-2',
'contains': 'bulkLattice/bravais'
}
},
'lat_row3': {
'parse_type': 'text',
'path_spec': {
'name': 'row-3',
'contains': 'bulkLattice/bravais'
}
},
'atom_positions': {
'parse_type': 'text',
'path_spec': {
'name': 'relPos'
}
},
'position_species': {
'parse_type': 'parentAttribs',
'path_spec': {
'name': 'relPos'
},
'flat': False,
'only_required': True
},
'element_species': {
'parse_type': 'allAttribs',
'path_spec': {
'name': 'species'
},
'flat': False,
'ignore': ['vcaAddCharge', 'magField']
},
},
'film_relax_info': {
'_general': True,
'_modes': [('relax', True), ('film', True)],
'_conversions': ['convert_relax_info'],
'lat_row1': {
'parse_type': 'text',
'path_spec': {
'name': 'row-1',
'contains': 'filmLattice/bravais'
}
},
'lat_row2': {
'parse_type': 'text',
'path_spec': {
'name': 'row-2',
'contains': 'filmLattice/bravais'
}
},
'lat_row3': {
'parse_type': 'text',
'path_spec': {
'name': 'row-3',
'contains': 'filmLattice/bravais'
}
},
'atom_positions': {
'parse_type': 'text',
'path_spec': {
'name': 'filmPos'
}
},
'position_species': {
'parse_type': 'parentAttribs',
'path_spec': {
'name': 'filmPos'
},
'flat': False,
'only_required': True
},
'element_species': {
'parse_type': 'allAttribs',
'path_spec': {
'name': 'species'
},
'flat': False,
'ignore': ['vcaAddCharge', 'magField']
},
},
#----General iteration tasks
# iteration number
# total energy (only total or also contributions, also lda+u correction)
# distances (nonmagnetic and magnetic, lda+u density matrix)
# charges (total, interstitial, mt sphere)
# fermi energy and bandgap
# magnetic moments
# orbital magnetic moments
# forces
'iteration_number': {
'_minimal': True,
'number_of_iterations_total': {
'parse_type': 'attrib',
'path_spec': {
'name': 'overallNumber'
},
'overwrite_last': True,
}
},
'total_energy': {
'_minimal': True,
'_conversions': ['convert_total_energy'],
'energy_hartree': {
'parse_type': 'singleValue',
'path_spec': {
'name': 'totalEnergy'
}
},
},
'distances': {
'_minimal': True,
'density_convergence': {
'parse_type': 'attrib',
'path_spec': {
'name': 'distance',
'tag_name': 'chargeDensity'
}
},
'density_convergence_units': {
'parse_type': 'attrib',
'path_spec': {
'name': 'units',
'tag_name': 'densityConvergence',
},
'overwrite_last': True,
}
},
'magnetic_distances': {
'_minimal': True,
'_modes': [('jspin', 2)],
'overall_density_convergence': {
'parse_type': 'attrib',
'path_spec': {
'name': 'distance',
'tag_name': 'overallChargeDensity'
}
},
'spin_density_convergence': {
'parse_type': 'attrib',
'path_spec': {
'name': 'distance',
'tag_name': 'spinDensity'
}
}
},
'total_energy_contributions': {
'sum_of_eigenvalues': {
'parse_type': 'singleValue',
'path_spec': {
'name': 'sumOfEigenvalues'
},
'only_required': True
},
'energy_core_electrons': {
'parse_type': 'singleValue',
'path_spec': {
'name': 'coreElectrons',
'contains': 'sumOfEigenvalues'
},
'only_required': True
},
'energy_valence_electrons': {
'parse_type': 'singleValue',
'path_spec': {
'name': 'valenceElectrons'
},
'only_required': True
},
'charge_den_xc_den_integral': {
'parse_type': 'singleValue',
'path_spec': {
'name': 'chargeDenXCDenIntegral'
},
'only_required': True
},
},
'ldau_energy_correction': {
'_modes': [('ldau', True)],
'ldau_energy_correction': {
'parse_type': 'singleValue',
'path_spec': {
'name': 'dftUCorrection'
},
'subdict': 'ldau_info',
'only_required': True
},
},
'nmmp_distances': {
'_minimal': True,
'_modes': [('ldau', True)],
'density_matrix_distance': {
'parse_type': 'attrib',
'path_spec': {
'name': 'distance',
'contains': 'ldaUDensityMatrixConvergence'
},
'subdict': 'ldau_info'
},
},
'fermi_energy': {
'fermi_energy': {
'parse_type': 'singleValue',
'path_spec': {
'name': 'FermiEnergy'
},
}
},
'bandgap': {
'_modes': [('bz_integration', 'hist')],
'bandgap': {
'parse_type': 'singleValue',
'path_spec': {
'name': 'bandgap'
},
}
},
'magnetic_moments': {
'_modes': [('jspin', 2)],
'magnetic_moments': {
'parse_type': 'allAttribs',
'path_spec': {
'name': 'magneticMoment'
},
'base_value': 'moment',
'ignore': ['atomType']
}
},
'orbital_magnetic_moments': {
'_modes': [('jspin', 2), ('soc', True)],
'orbital_magnetic_moments': {
'parse_type': 'allAttribs',
'path_spec': {
'name': 'orbMagMoment'
},
'base_value': 'moment',
'ignore': ['atomType']
}
},
'forces': {
'_minimal': True,
'_modes': [('relax', True)],
'_conversions': ['convert_forces'],
'force_units': {
'parse_type': 'attrib',
'path_spec': {
'name': 'units',
'tag_name': 'totalForcesOnRepresentativeAtoms'
},
'overwrite_last': True
},
'parsed_forces': {
'parse_type': 'allAttribs',
'path_spec': {
'name': 'forceTotal'
},
'flat': False,
'only_required': True
}
},
'charges': {
'_conversions': ['calculate_total_magnetic_moment'],
'spin_dependent_charge': {
'parse_type': 'allAttribs',
'path_spec': {
'name': 'spinDependentCharge',
'contains': 'allElectronCharges',
'not_contains': 'fixed'
},
'only_required': True
},
'total_charge': {
'parse_type': 'singleValue',
'path_spec': {
'name': 'totalCharge',
'contains': 'allElectronCharges',
'not_contains': 'fixed'
},
'only_required': True
}
},
#-------Tasks for forcetheorem Calculations
# DMI, JIJ, MAE, SSDISP
'forcetheorem_dmi': {
'_special': True,
'dmi_force': {
'parse_type': 'allAttribs',
'path_spec': {
'name': 'Entry',
'contains': 'DMI'
}
},
'dmi_force_qs': {
'parse_type': 'attrib',
'path_spec': {
'name': 'qpoints',
'contains': 'Forcetheorem_DMI'
}
},
'dmi_force_angles': {
'parse_type': 'attrib',
'path_spec': {
'name': 'Angles',
'contains': 'Forcetheorem_DMI'
}
},
'dmi_force_units': {
'parse_type': 'attrib',
'path_spec': {
'name': 'units',
'contains': 'Forcetheorem_DMI'
}
}
},
'forcetheorem_ssdisp': {
'_special': True,
'spst_force': {
'parse_type': 'allAttribs',
'path_spec': {
'name': 'Entry',
'contains': 'SSDISP'
}
},
'spst_force_qs': {
'parse_type': 'attrib',
'path_spec': {
'name': 'qvectors',
'contains': 'Forcetheorem_SSDISP'
}
},
'spst_force_units': {
'parse_type': 'attrib',
'path_spec': {
'name': 'units',
'contains': 'Forcetheorem_SSDISP'
}
}
},
'forcetheorem_mae': {
'_special': True,
'mae_force': {
'parse_type': 'allAttribs',
'path_spec': {
'name': 'Angle',
'contains': 'MAE'
}
},
'mae_force_units': {
'parse_type': 'attrib',
'path_spec': {
'name': 'units',
'contains': 'Forcetheorem_MAE'
}
}
},
'forcetheorem_jij': {
'_special': True,
'jij_force': {
'parse_type': 'allAttribs',
'path_spec': {
'name': 'Config',
'contains': 'JIJ'
}
},
'jij_force_units': {
'parse_type': 'attrib',
'path_spec': {
'name': 'units',
'contains': 'Forcetheorem_JIJ'
}
}
}
}
|
In this module migration functions for the task definitions are collected